
Listonella phage phiHSIC
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.27
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 47 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
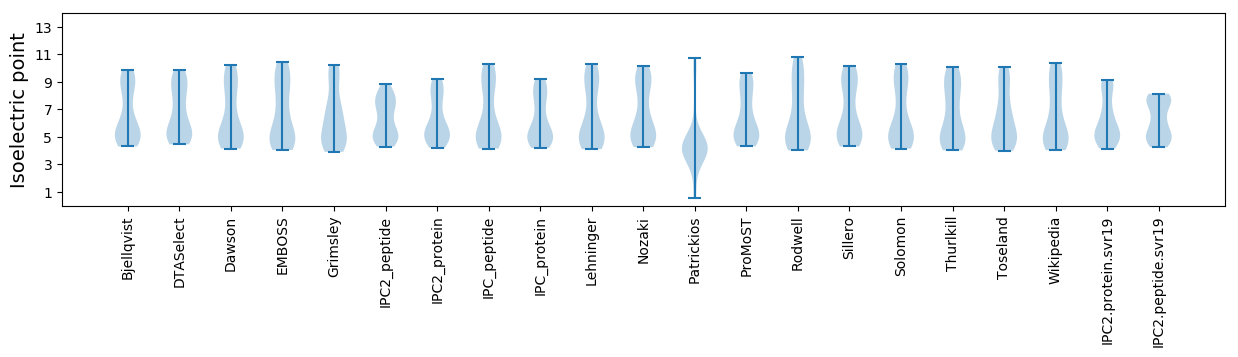
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q5G7N8|Q5G7N8_9CAUD Putative tail tape measure protein OS=Listonella phage phiHSIC OX=310539 GN=ORF31 PE=4 SV=1
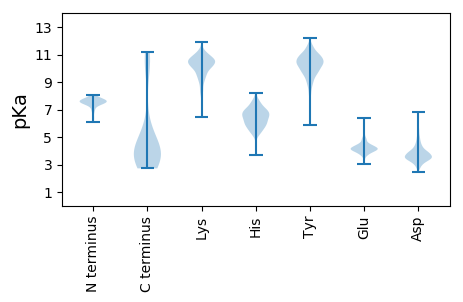
MM1 pKa = 7.19SLKK4 pKa = 10.47VVDD7 pKa = 5.05PIEE10 pKa = 3.98VTEE13 pKa = 4.26AVLTASGIPEE23 pKa = 4.31PDD25 pKa = 3.08VSQGEE30 pKa = 4.42VEE32 pKa = 4.35YY33 pKa = 11.22NPNKK37 pKa = 9.8TEE39 pKa = 4.56GIKK42 pKa = 9.25FTNVTWDD49 pKa = 2.95ITDD52 pKa = 3.57IARR55 pKa = 11.84DD56 pKa = 3.35GGTIYY61 pKa = 11.18ALDD64 pKa = 4.64EE65 pKa = 4.62YY66 pKa = 10.31PFSPSGASLVRR77 pKa = 11.84TYY79 pKa = 11.51DD80 pKa = 3.21VFGNEE85 pKa = 3.53TAIAFSTQNDD95 pKa = 3.77AKK97 pKa = 11.02CIDD100 pKa = 3.88VYY102 pKa = 11.06LGSIYY107 pKa = 11.06VGDD110 pKa = 3.73SNGFVNKK117 pKa = 10.16YY118 pKa = 6.62NTSGTLLEE126 pKa = 4.64TFDD129 pKa = 3.49LTANVSRR136 pKa = 11.84VDD138 pKa = 4.74AICGSYY144 pKa = 10.85GPFYY148 pKa = 10.66ILDD151 pKa = 3.55QTTGKK156 pKa = 9.95VFEE159 pKa = 4.91FGTLNDD165 pKa = 3.65LTPFEE170 pKa = 4.41VRR172 pKa = 11.84DD173 pKa = 4.0FGTFGSFIFGFTHH186 pKa = 6.76RR187 pKa = 11.84NGIIYY192 pKa = 7.42YY193 pKa = 8.73TNQNNGKK200 pKa = 8.63IEE202 pKa = 4.23SMSAGTGEE210 pKa = 4.32ARR212 pKa = 11.84DD213 pKa = 4.01SFALTNDD220 pKa = 3.31RR221 pKa = 11.84DD222 pKa = 3.66GVYY225 pKa = 10.77YY226 pKa = 10.91LGGLAFLADD235 pKa = 3.96GSLLSVNTTGSEE247 pKa = 3.7AFKK250 pKa = 10.86YY251 pKa = 10.89VNGSLKK257 pKa = 10.29QPYY260 pKa = 9.7NPGDD264 pKa = 3.58QVVVLSNHH272 pKa = 5.65TKK274 pKa = 9.04YY275 pKa = 10.29QCLVATSQSPIDD287 pKa = 4.28GATEE291 pKa = 3.92DD292 pKa = 5.01ATATWIKK299 pKa = 10.63VGPTNKK305 pKa = 9.01WAMFDD310 pKa = 3.47NLQNTKK316 pKa = 8.39TLNDD320 pKa = 3.3TDD322 pKa = 4.05FTITLNPTTYY332 pKa = 10.85VNTLAALGFSGVTSIRR348 pKa = 11.84VEE350 pKa = 3.77VDD352 pKa = 2.88NVGGTTIYY360 pKa = 10.94DD361 pKa = 3.64KK362 pKa = 11.36TFGTSDD368 pKa = 3.42FSAIYY373 pKa = 10.4DD374 pKa = 3.39HH375 pKa = 6.09YY376 pKa = 10.06TYY378 pKa = 11.36VFYY381 pKa = 10.9QIVALQKK388 pKa = 10.88LIVDD392 pKa = 5.03DD393 pKa = 5.52LPPLPDD399 pKa = 3.21TEE401 pKa = 4.08IRR403 pKa = 11.84VTFSGSDD410 pKa = 3.22IQIGEE415 pKa = 4.37LAHH418 pKa = 6.68GFAINVGTLVAEE430 pKa = 4.39STKK433 pKa = 10.51SDD435 pKa = 3.04RR436 pKa = 11.84FRR438 pKa = 11.84YY439 pKa = 9.69RR440 pKa = 11.84EE441 pKa = 3.59QQYY444 pKa = 10.89NEE446 pKa = 3.55FGYY449 pKa = 9.9PVGAEE454 pKa = 4.15PIVVEE459 pKa = 4.23LNTYY463 pKa = 10.43DD464 pKa = 3.81VLVPKK469 pKa = 10.43LNNPAIQKK477 pKa = 10.41LLDD480 pKa = 3.66RR481 pKa = 11.84LTGKK485 pKa = 8.23NTLWVGDD492 pKa = 3.53IGGGQGLITYY502 pKa = 8.02GFFEE506 pKa = 4.79RR507 pKa = 11.84SPIPFSMPYY516 pKa = 10.0DD517 pKa = 3.28INYY520 pKa = 9.7QITVRR525 pKa = 11.84ASVV528 pKa = 2.9
MM1 pKa = 7.19SLKK4 pKa = 10.47VVDD7 pKa = 5.05PIEE10 pKa = 3.98VTEE13 pKa = 4.26AVLTASGIPEE23 pKa = 4.31PDD25 pKa = 3.08VSQGEE30 pKa = 4.42VEE32 pKa = 4.35YY33 pKa = 11.22NPNKK37 pKa = 9.8TEE39 pKa = 4.56GIKK42 pKa = 9.25FTNVTWDD49 pKa = 2.95ITDD52 pKa = 3.57IARR55 pKa = 11.84DD56 pKa = 3.35GGTIYY61 pKa = 11.18ALDD64 pKa = 4.64EE65 pKa = 4.62YY66 pKa = 10.31PFSPSGASLVRR77 pKa = 11.84TYY79 pKa = 11.51DD80 pKa = 3.21VFGNEE85 pKa = 3.53TAIAFSTQNDD95 pKa = 3.77AKK97 pKa = 11.02CIDD100 pKa = 3.88VYY102 pKa = 11.06LGSIYY107 pKa = 11.06VGDD110 pKa = 3.73SNGFVNKK117 pKa = 10.16YY118 pKa = 6.62NTSGTLLEE126 pKa = 4.64TFDD129 pKa = 3.49LTANVSRR136 pKa = 11.84VDD138 pKa = 4.74AICGSYY144 pKa = 10.85GPFYY148 pKa = 10.66ILDD151 pKa = 3.55QTTGKK156 pKa = 9.95VFEE159 pKa = 4.91FGTLNDD165 pKa = 3.65LTPFEE170 pKa = 4.41VRR172 pKa = 11.84DD173 pKa = 4.0FGTFGSFIFGFTHH186 pKa = 6.76RR187 pKa = 11.84NGIIYY192 pKa = 7.42YY193 pKa = 8.73TNQNNGKK200 pKa = 8.63IEE202 pKa = 4.23SMSAGTGEE210 pKa = 4.32ARR212 pKa = 11.84DD213 pKa = 4.01SFALTNDD220 pKa = 3.31RR221 pKa = 11.84DD222 pKa = 3.66GVYY225 pKa = 10.77YY226 pKa = 10.91LGGLAFLADD235 pKa = 3.96GSLLSVNTTGSEE247 pKa = 3.7AFKK250 pKa = 10.86YY251 pKa = 10.89VNGSLKK257 pKa = 10.29QPYY260 pKa = 9.7NPGDD264 pKa = 3.58QVVVLSNHH272 pKa = 5.65TKK274 pKa = 9.04YY275 pKa = 10.29QCLVATSQSPIDD287 pKa = 4.28GATEE291 pKa = 3.92DD292 pKa = 5.01ATATWIKK299 pKa = 10.63VGPTNKK305 pKa = 9.01WAMFDD310 pKa = 3.47NLQNTKK316 pKa = 8.39TLNDD320 pKa = 3.3TDD322 pKa = 4.05FTITLNPTTYY332 pKa = 10.85VNTLAALGFSGVTSIRR348 pKa = 11.84VEE350 pKa = 3.77VDD352 pKa = 2.88NVGGTTIYY360 pKa = 10.94DD361 pKa = 3.64KK362 pKa = 11.36TFGTSDD368 pKa = 3.42FSAIYY373 pKa = 10.4DD374 pKa = 3.39HH375 pKa = 6.09YY376 pKa = 10.06TYY378 pKa = 11.36VFYY381 pKa = 10.9QIVALQKK388 pKa = 10.88LIVDD392 pKa = 5.03DD393 pKa = 5.52LPPLPDD399 pKa = 3.21TEE401 pKa = 4.08IRR403 pKa = 11.84VTFSGSDD410 pKa = 3.22IQIGEE415 pKa = 4.37LAHH418 pKa = 6.68GFAINVGTLVAEE430 pKa = 4.39STKK433 pKa = 10.51SDD435 pKa = 3.04RR436 pKa = 11.84FRR438 pKa = 11.84YY439 pKa = 9.69RR440 pKa = 11.84EE441 pKa = 3.59QQYY444 pKa = 10.89NEE446 pKa = 3.55FGYY449 pKa = 9.9PVGAEE454 pKa = 4.15PIVVEE459 pKa = 4.23LNTYY463 pKa = 10.43DD464 pKa = 3.81VLVPKK469 pKa = 10.43LNNPAIQKK477 pKa = 10.41LLDD480 pKa = 3.66RR481 pKa = 11.84LTGKK485 pKa = 8.23NTLWVGDD492 pKa = 3.53IGGGQGLITYY502 pKa = 8.02GFFEE506 pKa = 4.79RR507 pKa = 11.84SPIPFSMPYY516 pKa = 10.0DD517 pKa = 3.28INYY520 pKa = 9.7QITVRR525 pKa = 11.84ASVV528 pKa = 2.9
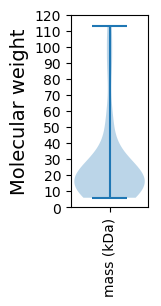
Molecular weight: 57.81 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q5G7P5|Q5G7P5_9CAUD Hypothetical phage protein OS=Listonella phage phiHSIC OX=310539 GN=ORF24 PE=4 SV=1
MM1 pKa = 7.63LSCICGGNPMSWSKK15 pKa = 10.79SLKK18 pKa = 10.42NIIVKK23 pKa = 10.4NEE25 pKa = 3.65NLTEE29 pKa = 4.04KK30 pKa = 10.41QLRR33 pKa = 11.84AGLFDD38 pKa = 4.03AANTVILGSPVGAPEE53 pKa = 5.09LWQQPAPNYY62 pKa = 9.72YY63 pKa = 9.66RR64 pKa = 11.84AGSYY68 pKa = 10.16RR69 pKa = 11.84SNHH72 pKa = 5.7RR73 pKa = 11.84VSISKK78 pKa = 8.29ITSFEE83 pKa = 4.06KK84 pKa = 10.87GISSQSSIMMDD95 pKa = 4.02LQSDD99 pKa = 3.55IAKK102 pKa = 10.16FKK104 pKa = 10.6IGEE107 pKa = 4.39TLFMTNPLPYY117 pKa = 9.03ATSIEE122 pKa = 4.36YY123 pKa = 10.21GHH125 pKa = 6.67SSQAPNGVYY134 pKa = 9.86RR135 pKa = 11.84PAVRR139 pKa = 11.84RR140 pKa = 11.84LVKK143 pKa = 10.39FLNTEE148 pKa = 4.55LKK150 pKa = 10.81AKK152 pKa = 10.3
MM1 pKa = 7.63LSCICGGNPMSWSKK15 pKa = 10.79SLKK18 pKa = 10.42NIIVKK23 pKa = 10.4NEE25 pKa = 3.65NLTEE29 pKa = 4.04KK30 pKa = 10.41QLRR33 pKa = 11.84AGLFDD38 pKa = 4.03AANTVILGSPVGAPEE53 pKa = 5.09LWQQPAPNYY62 pKa = 9.72YY63 pKa = 9.66RR64 pKa = 11.84AGSYY68 pKa = 10.16RR69 pKa = 11.84SNHH72 pKa = 5.7RR73 pKa = 11.84VSISKK78 pKa = 8.29ITSFEE83 pKa = 4.06KK84 pKa = 10.87GISSQSSIMMDD95 pKa = 4.02LQSDD99 pKa = 3.55IAKK102 pKa = 10.16FKK104 pKa = 10.6IGEE107 pKa = 4.39TLFMTNPLPYY117 pKa = 9.03ATSIEE122 pKa = 4.36YY123 pKa = 10.21GHH125 pKa = 6.67SSQAPNGVYY134 pKa = 9.86RR135 pKa = 11.84PAVRR139 pKa = 11.84RR140 pKa = 11.84LVKK143 pKa = 10.39FLNTEE148 pKa = 4.55LKK150 pKa = 10.81AKK152 pKa = 10.3
Molecular weight: 16.82 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
11099 |
49 |
1069 |
236.1 |
26.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.226 ± 0.608 | 1.505 ± 0.269 |
6.118 ± 0.236 | 6.92 ± 0.372 |
4.036 ± 0.181 | 7.028 ± 0.349 |
1.739 ± 0.223 | 5.928 ± 0.239 |
6.902 ± 0.435 | 7.947 ± 0.354 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.874 ± 0.209 | 5.28 ± 0.257 |
3.559 ± 0.256 | 3.721 ± 0.284 |
3.982 ± 0.25 | 6.965 ± 0.388 |
5.739 ± 0.364 | 6.631 ± 0.257 |
1.189 ± 0.159 | 3.694 ± 0.253 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
