
Candidatus Aquiluna sp. 15G-AUS-rot
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Luna cluster; Luna-1 subcluster; Aquiluna
Average proteome isoelectric point is 6.14
Get precalculated fractions of proteins
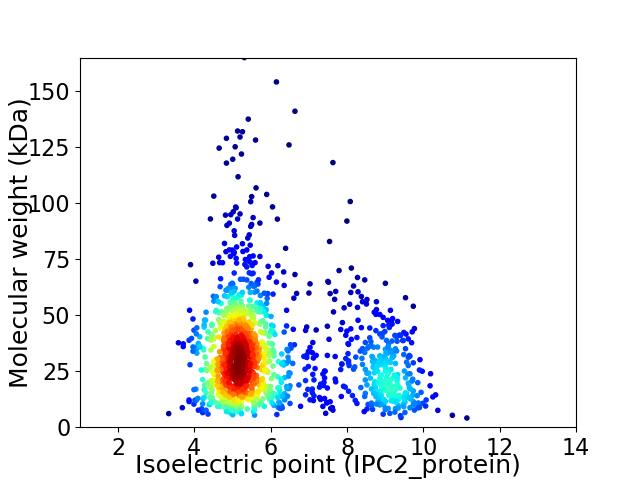
Virtual 2D-PAGE plot for 1370 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A7D4U7R3|A0A7D4U7R3_9MICO Tryptophan synthase beta chain OS=Candidatus Aquiluna sp. 15G-AUS-rot OX=2499157 GN=trpB PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 10.39LKK4 pKa = 10.39NASAAFALLGASALVLSGCAAAPEE28 pKa = 4.35AEE30 pKa = 4.26PTATEE35 pKa = 4.33SEE37 pKa = 4.63TATEE41 pKa = 3.91EE42 pKa = 3.94AAPSVDD48 pKa = 3.61YY49 pKa = 10.26LACAVSDD56 pKa = 3.51EE57 pKa = 4.7GSWNDD62 pKa = 2.93KK63 pKa = 10.71SFNEE67 pKa = 4.19SVYY70 pKa = 11.22DD71 pKa = 3.8GLLKK75 pKa = 10.78AEE77 pKa = 4.81AEE79 pKa = 4.29LGVQIAEE86 pKa = 4.33AEE88 pKa = 4.5SNSAEE93 pKa = 4.08DD94 pKa = 4.04FAPNLQAMVDD104 pKa = 3.75QACDD108 pKa = 2.77ITFAVGFNLVADD120 pKa = 4.31VNAAAAANPEE130 pKa = 4.38TQFVTIDD137 pKa = 2.97GWSEE141 pKa = 3.61GNANLKK147 pKa = 9.42PVGYY151 pKa = 11.13AMNQSSYY158 pKa = 10.83LAGYY162 pKa = 8.74LAAAYY167 pKa = 7.6STTKK171 pKa = 10.55VVGTYY176 pKa = 10.99GGMQIDD182 pKa = 3.91AVTDD186 pKa = 3.82FMNGFYY192 pKa = 10.18YY193 pKa = 10.86GAMAWGAEE201 pKa = 3.88NGAEE205 pKa = 4.11VKK207 pKa = 10.61VVGWDD212 pKa = 3.28PAAATGQFIGDD223 pKa = 4.06FTPNSGTSKK232 pKa = 10.64SIAAALINDD241 pKa = 4.16GADD244 pKa = 3.07VIFPVGGDD252 pKa = 3.09QFGAVSQAIEE262 pKa = 3.83EE263 pKa = 4.14AGIAGVMIGVDD274 pKa = 3.29KK275 pKa = 11.08DD276 pKa = 3.67VALTSPEE283 pKa = 3.86YY284 pKa = 10.7APYY287 pKa = 11.12VLTSAEE293 pKa = 3.86KK294 pKa = 10.26RR295 pKa = 11.84MTNAVYY301 pKa = 10.45DD302 pKa = 3.96IVAEE306 pKa = 4.35LSAGGEE312 pKa = 4.23FSGDD316 pKa = 3.24AYY318 pKa = 11.22LGTLANGGTGLSPFYY333 pKa = 10.74EE334 pKa = 4.55FDD336 pKa = 3.98SKK338 pKa = 11.17IDD340 pKa = 3.77DD341 pKa = 3.55ATKK344 pKa = 10.74ARR346 pKa = 11.84LAEE349 pKa = 4.19LEE351 pKa = 3.95AGIIAGEE358 pKa = 4.01IDD360 pKa = 3.75PLSS363 pKa = 3.6
MM1 pKa = 7.45KK2 pKa = 10.39LKK4 pKa = 10.39NASAAFALLGASALVLSGCAAAPEE28 pKa = 4.35AEE30 pKa = 4.26PTATEE35 pKa = 4.33SEE37 pKa = 4.63TATEE41 pKa = 3.91EE42 pKa = 3.94AAPSVDD48 pKa = 3.61YY49 pKa = 10.26LACAVSDD56 pKa = 3.51EE57 pKa = 4.7GSWNDD62 pKa = 2.93KK63 pKa = 10.71SFNEE67 pKa = 4.19SVYY70 pKa = 11.22DD71 pKa = 3.8GLLKK75 pKa = 10.78AEE77 pKa = 4.81AEE79 pKa = 4.29LGVQIAEE86 pKa = 4.33AEE88 pKa = 4.5SNSAEE93 pKa = 4.08DD94 pKa = 4.04FAPNLQAMVDD104 pKa = 3.75QACDD108 pKa = 2.77ITFAVGFNLVADD120 pKa = 4.31VNAAAAANPEE130 pKa = 4.38TQFVTIDD137 pKa = 2.97GWSEE141 pKa = 3.61GNANLKK147 pKa = 9.42PVGYY151 pKa = 11.13AMNQSSYY158 pKa = 10.83LAGYY162 pKa = 8.74LAAAYY167 pKa = 7.6STTKK171 pKa = 10.55VVGTYY176 pKa = 10.99GGMQIDD182 pKa = 3.91AVTDD186 pKa = 3.82FMNGFYY192 pKa = 10.18YY193 pKa = 10.86GAMAWGAEE201 pKa = 3.88NGAEE205 pKa = 4.11VKK207 pKa = 10.61VVGWDD212 pKa = 3.28PAAATGQFIGDD223 pKa = 4.06FTPNSGTSKK232 pKa = 10.64SIAAALINDD241 pKa = 4.16GADD244 pKa = 3.07VIFPVGGDD252 pKa = 3.09QFGAVSQAIEE262 pKa = 3.83EE263 pKa = 4.14AGIAGVMIGVDD274 pKa = 3.29KK275 pKa = 11.08DD276 pKa = 3.67VALTSPEE283 pKa = 3.86YY284 pKa = 10.7APYY287 pKa = 11.12VLTSAEE293 pKa = 3.86KK294 pKa = 10.26RR295 pKa = 11.84MTNAVYY301 pKa = 10.45DD302 pKa = 3.96IVAEE306 pKa = 4.35LSAGGEE312 pKa = 4.23FSGDD316 pKa = 3.24AYY318 pKa = 11.22LGTLANGGTGLSPFYY333 pKa = 10.74EE334 pKa = 4.55FDD336 pKa = 3.98SKK338 pKa = 11.17IDD340 pKa = 3.77DD341 pKa = 3.55ATKK344 pKa = 10.74ARR346 pKa = 11.84LAEE349 pKa = 4.19LEE351 pKa = 3.95AGIIAGEE358 pKa = 4.01IDD360 pKa = 3.75PLSS363 pKa = 3.6
Molecular weight: 37.26 kDa
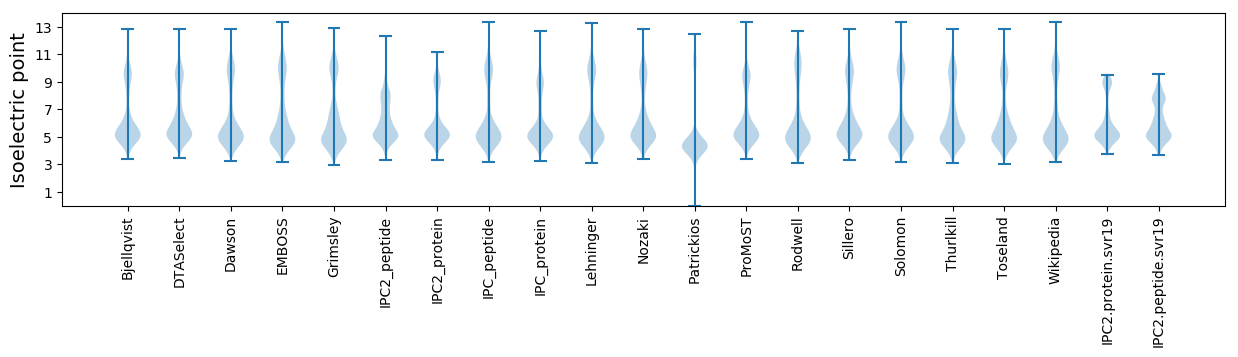
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A7D4PZX1|A0A7D4PZX1_9MICO Aminobutyraldehyde dehydrogenase OS=Candidatus Aquiluna sp. 15G-AUS-rot OX=2499157 GN=HRU87_04745 PE=3 SV=1
MM1 pKa = 7.46GSIIKK6 pKa = 9.98KK7 pKa = 8.35RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.39RR11 pKa = 11.84MSKK14 pKa = 9.76KK15 pKa = 9.54KK16 pKa = 9.79HH17 pKa = 5.67RR18 pKa = 11.84KK19 pKa = 8.31LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.46GSIIKK6 pKa = 9.98KK7 pKa = 8.35RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.39RR11 pKa = 11.84MSKK14 pKa = 9.76KK15 pKa = 9.54KK16 pKa = 9.79HH17 pKa = 5.67RR18 pKa = 11.84KK19 pKa = 8.31LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
429841 |
32 |
1516 |
313.8 |
34.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.844 ± 0.085 | 0.528 ± 0.014 |
5.125 ± 0.048 | 6.574 ± 0.065 |
3.895 ± 0.044 | 8.1 ± 0.047 |
1.776 ± 0.032 | 5.677 ± 0.045 |
4.217 ± 0.05 | 10.785 ± 0.08 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.084 ± 0.024 | 3.103 ± 0.038 |
4.517 ± 0.034 | 3.667 ± 0.033 |
5.506 ± 0.052 | 6.796 ± 0.052 |
5.25 ± 0.061 | 7.996 ± 0.055 |
1.296 ± 0.027 | 2.265 ± 0.035 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
