
Frigoribacterium sp. Leaf164
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Frigoribacterium; unclassified Frigoribacterium
Average proteome isoelectric point is 5.96
Get precalculated fractions of proteins
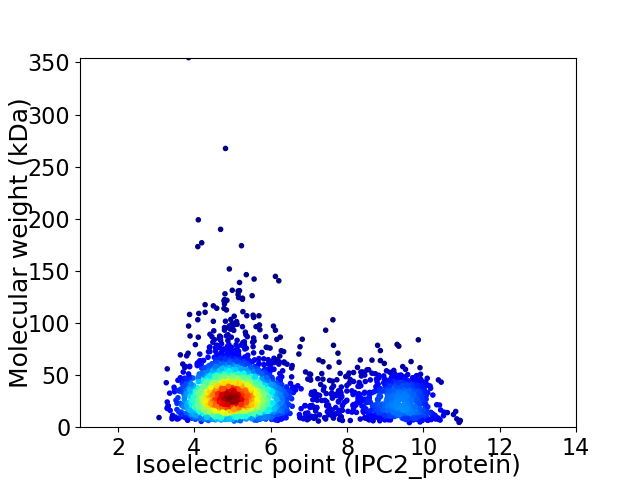
Virtual 2D-PAGE plot for 2793 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Q5LKL0|A0A0Q5LKL0_9MICO Ribonuclease P protein component OS=Frigoribacterium sp. Leaf164 OX=1736282 GN=rnpA PE=3 SV=1
MM1 pKa = 7.35GVLIIAASAAGIAAFGASTAPAPGARR27 pKa = 11.84LTPSEE32 pKa = 4.12QGEE35 pKa = 4.53TTVTVRR41 pKa = 11.84LWDD44 pKa = 3.45EE45 pKa = 4.14TVAAGYY51 pKa = 7.69EE52 pKa = 3.92KK53 pKa = 10.82SFAAFEE59 pKa = 4.37RR60 pKa = 11.84DD61 pKa = 3.49NPDD64 pKa = 2.47IDD66 pKa = 4.21VEE68 pKa = 4.67VEE70 pKa = 4.0VVPYY74 pKa = 10.95ADD76 pKa = 5.05YY77 pKa = 10.92FDD79 pKa = 4.6QLADD83 pKa = 4.41DD84 pKa = 4.77VDD86 pKa = 5.95DD87 pKa = 5.36GDD89 pKa = 4.97APDD92 pKa = 4.7VYY94 pKa = 10.36WLNSSHH100 pKa = 6.54YY101 pKa = 10.16DD102 pKa = 3.52GYY104 pKa = 11.76AEE106 pKa = 5.42AGDD109 pKa = 4.39LVDD112 pKa = 3.86VDD114 pKa = 4.4AVLGEE119 pKa = 4.17SARR122 pKa = 11.84RR123 pKa = 11.84AWSPAVVDD131 pKa = 3.89QFTHH135 pKa = 7.35DD136 pKa = 3.41GSLWGVPQLSDD147 pKa = 3.34GGIALYY153 pKa = 11.11YY154 pKa = 10.66NADD157 pKa = 3.73LLAAAGVDD165 pKa = 3.75PASLDD170 pKa = 3.75DD171 pKa = 4.28LTWAPGGGADD181 pKa = 5.03DD182 pKa = 3.96TLLPLLQRR190 pKa = 11.84LTLDD194 pKa = 3.58DD195 pKa = 3.7QGRR198 pKa = 11.84NAADD202 pKa = 3.45PAFDD206 pKa = 3.74GTVTQYY212 pKa = 11.36GYY214 pKa = 11.11NAGNDD219 pKa = 3.92LQAIWYY225 pKa = 8.09DD226 pKa = 3.8YY227 pKa = 10.95LGSAGGRR234 pKa = 11.84LEE236 pKa = 4.6DD237 pKa = 3.85PEE239 pKa = 4.5GASFEE244 pKa = 4.86LDD246 pKa = 3.3TPEE249 pKa = 3.95AQAAFQYY256 pKa = 10.94LVDD259 pKa = 4.05LVGVRR264 pKa = 11.84RR265 pKa = 11.84VAPPAAATNADD276 pKa = 3.92PDD278 pKa = 3.45HH279 pKa = 6.86SRR281 pKa = 11.84DD282 pKa = 3.67LFTAGHH288 pKa = 5.94MALFQSGLYY297 pKa = 9.85SLSTIEE303 pKa = 4.87SQAQFDD309 pKa = 3.89WGVAMMPAGPAGRR322 pKa = 11.84VSVSNGIVAAGDD334 pKa = 3.21AHH336 pKa = 6.15TAHH339 pKa = 7.35LDD341 pKa = 3.4DD342 pKa = 4.11TAKK345 pKa = 10.54VLAWLGSTDD354 pKa = 3.02GAEE357 pKa = 4.2YY358 pKa = 10.11IGRR361 pKa = 11.84DD362 pKa = 3.57GAAVPAVTGAQEE374 pKa = 3.64PYY376 pKa = 7.84YY377 pKa = 10.51AYY379 pKa = 8.86WASRR383 pKa = 11.84GVDD386 pKa = 3.03VSPFFDD392 pKa = 4.25VIASGDD398 pKa = 4.01TIPAPTSLYY407 pKa = 9.49GTAQPALDD415 pKa = 4.37PLLDD419 pKa = 3.61QVFAGSLPVPEE430 pKa = 4.92GLAAAQAAAEE440 pKa = 4.18AAVAAAEE447 pKa = 4.13QASDD451 pKa = 4.0PSATPAPP458 pKa = 4.07
MM1 pKa = 7.35GVLIIAASAAGIAAFGASTAPAPGARR27 pKa = 11.84LTPSEE32 pKa = 4.12QGEE35 pKa = 4.53TTVTVRR41 pKa = 11.84LWDD44 pKa = 3.45EE45 pKa = 4.14TVAAGYY51 pKa = 7.69EE52 pKa = 3.92KK53 pKa = 10.82SFAAFEE59 pKa = 4.37RR60 pKa = 11.84DD61 pKa = 3.49NPDD64 pKa = 2.47IDD66 pKa = 4.21VEE68 pKa = 4.67VEE70 pKa = 4.0VVPYY74 pKa = 10.95ADD76 pKa = 5.05YY77 pKa = 10.92FDD79 pKa = 4.6QLADD83 pKa = 4.41DD84 pKa = 4.77VDD86 pKa = 5.95DD87 pKa = 5.36GDD89 pKa = 4.97APDD92 pKa = 4.7VYY94 pKa = 10.36WLNSSHH100 pKa = 6.54YY101 pKa = 10.16DD102 pKa = 3.52GYY104 pKa = 11.76AEE106 pKa = 5.42AGDD109 pKa = 4.39LVDD112 pKa = 3.86VDD114 pKa = 4.4AVLGEE119 pKa = 4.17SARR122 pKa = 11.84RR123 pKa = 11.84AWSPAVVDD131 pKa = 3.89QFTHH135 pKa = 7.35DD136 pKa = 3.41GSLWGVPQLSDD147 pKa = 3.34GGIALYY153 pKa = 11.11YY154 pKa = 10.66NADD157 pKa = 3.73LLAAAGVDD165 pKa = 3.75PASLDD170 pKa = 3.75DD171 pKa = 4.28LTWAPGGGADD181 pKa = 5.03DD182 pKa = 3.96TLLPLLQRR190 pKa = 11.84LTLDD194 pKa = 3.58DD195 pKa = 3.7QGRR198 pKa = 11.84NAADD202 pKa = 3.45PAFDD206 pKa = 3.74GTVTQYY212 pKa = 11.36GYY214 pKa = 11.11NAGNDD219 pKa = 3.92LQAIWYY225 pKa = 8.09DD226 pKa = 3.8YY227 pKa = 10.95LGSAGGRR234 pKa = 11.84LEE236 pKa = 4.6DD237 pKa = 3.85PEE239 pKa = 4.5GASFEE244 pKa = 4.86LDD246 pKa = 3.3TPEE249 pKa = 3.95AQAAFQYY256 pKa = 10.94LVDD259 pKa = 4.05LVGVRR264 pKa = 11.84RR265 pKa = 11.84VAPPAAATNADD276 pKa = 3.92PDD278 pKa = 3.45HH279 pKa = 6.86SRR281 pKa = 11.84DD282 pKa = 3.67LFTAGHH288 pKa = 5.94MALFQSGLYY297 pKa = 9.85SLSTIEE303 pKa = 4.87SQAQFDD309 pKa = 3.89WGVAMMPAGPAGRR322 pKa = 11.84VSVSNGIVAAGDD334 pKa = 3.21AHH336 pKa = 6.15TAHH339 pKa = 7.35LDD341 pKa = 3.4DD342 pKa = 4.11TAKK345 pKa = 10.54VLAWLGSTDD354 pKa = 3.02GAEE357 pKa = 4.2YY358 pKa = 10.11IGRR361 pKa = 11.84DD362 pKa = 3.57GAAVPAVTGAQEE374 pKa = 3.64PYY376 pKa = 7.84YY377 pKa = 10.51AYY379 pKa = 8.86WASRR383 pKa = 11.84GVDD386 pKa = 3.03VSPFFDD392 pKa = 4.25VIASGDD398 pKa = 4.01TIPAPTSLYY407 pKa = 9.49GTAQPALDD415 pKa = 4.37PLLDD419 pKa = 3.61QVFAGSLPVPEE430 pKa = 4.92GLAAAQAAAEE440 pKa = 4.18AAVAAAEE447 pKa = 4.13QASDD451 pKa = 4.0PSATPAPP458 pKa = 4.07
Molecular weight: 47.44 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q5LH32|A0A0Q5LH32_9MICO Peptide ABC transporter ATP-binding protein OS=Frigoribacterium sp. Leaf164 OX=1736282 GN=ASF82_14635 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 10.04KK16 pKa = 8.86HH17 pKa = 4.36GFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILAARR35 pKa = 11.84RR36 pKa = 11.84GKK38 pKa = 10.68GRR40 pKa = 11.84TEE42 pKa = 4.13LSAA45 pKa = 4.86
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 10.04KK16 pKa = 8.86HH17 pKa = 4.36GFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILAARR35 pKa = 11.84RR36 pKa = 11.84GKK38 pKa = 10.68GRR40 pKa = 11.84TEE42 pKa = 4.13LSAA45 pKa = 4.86
Molecular weight: 5.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
897636 |
38 |
3668 |
321.4 |
34.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.947 ± 0.066 | 0.419 ± 0.01 |
6.559 ± 0.044 | 5.372 ± 0.049 |
2.832 ± 0.029 | 9.399 ± 0.047 |
1.903 ± 0.024 | 3.193 ± 0.042 |
1.653 ± 0.032 | 10.173 ± 0.059 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.503 ± 0.017 | 1.508 ± 0.022 |
5.556 ± 0.037 | 2.681 ± 0.028 |
7.337 ± 0.054 | 5.909 ± 0.034 |
6.391 ± 0.061 | 10.466 ± 0.062 |
1.434 ± 0.019 | 1.764 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
