
Alces alces faeces associated microvirus MP3 6497
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.09
Get precalculated fractions of proteins
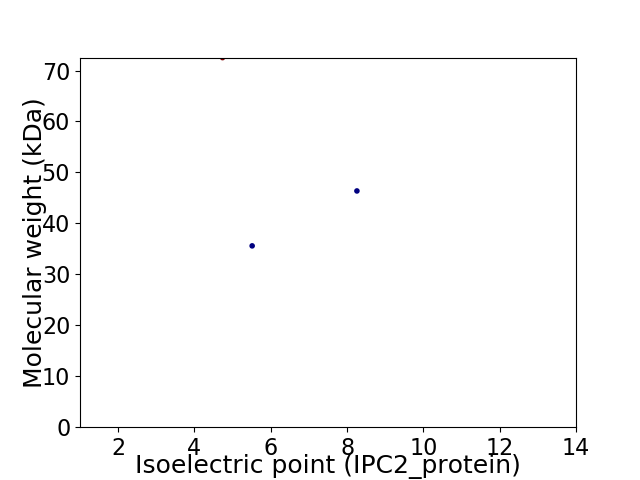
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
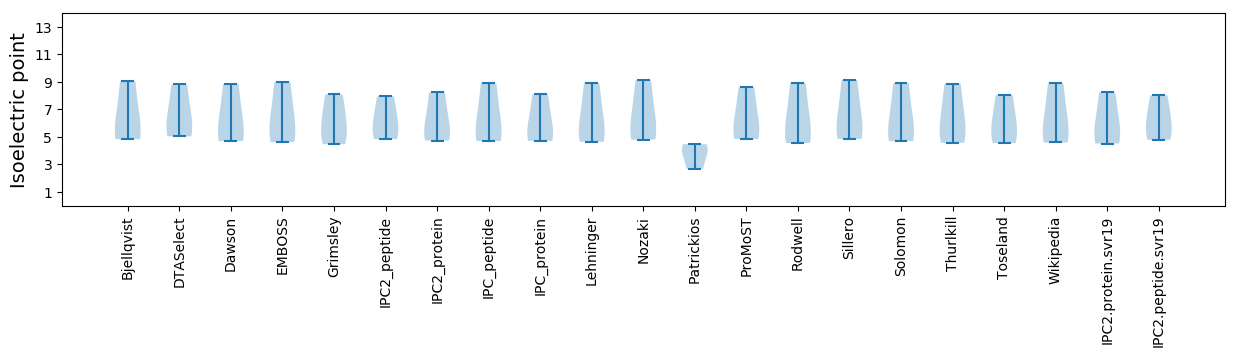
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2Z5CHK4|A0A2Z5CHK4_9VIRU Minor capsid protein OS=Alces alces faeces associated microvirus MP3 6497 OX=2219139 PE=4 SV=1
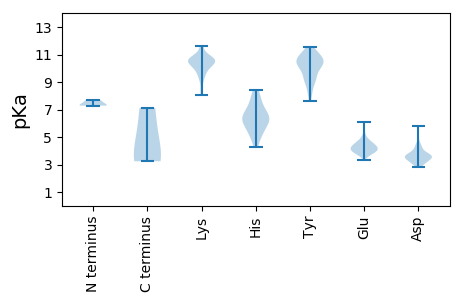
MM1 pKa = 7.68ANRR4 pKa = 11.84YY5 pKa = 9.16DD6 pKa = 4.22RR7 pKa = 11.84LDD9 pKa = 3.22NTAPIEE15 pKa = 4.21TALKK19 pKa = 9.98ADD21 pKa = 3.76SVSVGRR27 pKa = 11.84SAFDD31 pKa = 3.08WSCIHH36 pKa = 5.9NTNGIIGSLIPIDD49 pKa = 4.31CFDD52 pKa = 3.72VVPNEE57 pKa = 4.66DD58 pKa = 4.0LDD60 pKa = 3.74ISIASLIEE68 pKa = 3.79FRR70 pKa = 11.84NPTTRR75 pKa = 11.84QLFNGMRR82 pKa = 11.84VFVHH86 pKa = 7.01AYY88 pKa = 8.93YY89 pKa = 10.9NRR91 pKa = 11.84LTDD94 pKa = 3.22LWEE97 pKa = 4.52GARR100 pKa = 11.84NWLDD104 pKa = 3.12NGRR107 pKa = 11.84TGKK110 pKa = 10.75LSLTRR115 pKa = 11.84PNLVYY120 pKa = 10.41TGFYY124 pKa = 9.62KK125 pKa = 10.77APSNVTYY132 pKa = 10.47RR133 pKa = 11.84GNACTPMSLLDD144 pKa = 3.96YY145 pKa = 10.76LGLPAQRR152 pKa = 11.84MQDD155 pKa = 3.38TLKK158 pKa = 10.2TSDD161 pKa = 3.47GSVTASPLRR170 pKa = 11.84SFQCAYY176 pKa = 9.4WSSNNDD182 pKa = 2.99FTASEE187 pKa = 4.31LSYY190 pKa = 11.49APDD193 pKa = 4.54FFPADD198 pKa = 3.17CCMAYY203 pKa = 9.78QRR205 pKa = 11.84NWRR208 pKa = 11.84DD209 pKa = 3.53FYY211 pKa = 11.56CNKK214 pKa = 10.14NLLQNNPYY222 pKa = 9.12WFPEE226 pKa = 4.06NEE228 pKa = 3.75EE229 pKa = 4.26HH230 pKa = 7.71FILSYY235 pKa = 10.3SCSDD239 pKa = 3.55AVCVNYY245 pKa = 10.65GNEE248 pKa = 4.07NLSSVSANALNNIVGNIPTVIADD271 pKa = 3.46YY272 pKa = 11.19SVTPEE277 pKa = 4.09PNNPITNANDD287 pKa = 3.37YY288 pKa = 10.17PGYY291 pKa = 10.23IPTYY295 pKa = 10.12APNLSGLKK303 pKa = 9.72FRR305 pKa = 11.84QFRR308 pKa = 11.84GDD310 pKa = 3.22RR311 pKa = 11.84FTTALPFPDD320 pKa = 5.8LIRR323 pKa = 11.84GDD325 pKa = 3.89IPILEE330 pKa = 4.24YY331 pKa = 10.71TSDD334 pKa = 3.45NVASVTFTRR343 pKa = 11.84SDD345 pKa = 3.64DD346 pKa = 3.94DD347 pKa = 4.27SVEE350 pKa = 4.28LDD352 pKa = 3.18LHH354 pKa = 6.43NNYY357 pKa = 10.34SSTIGGTYY365 pKa = 10.12VNTYY369 pKa = 10.37LKK371 pKa = 10.77DD372 pKa = 3.58STVEE376 pKa = 3.78NTDD379 pKa = 3.53PFNVTATVDD388 pKa = 3.88LSSLGITMSDD398 pKa = 2.92IYY400 pKa = 10.96TLEE403 pKa = 3.83TLTAFRR409 pKa = 11.84RR410 pKa = 11.84RR411 pKa = 11.84MGMTNGDD418 pKa = 4.04YY419 pKa = 11.51NEE421 pKa = 4.62MIKK424 pKa = 10.49AQYY427 pKa = 9.32GVNPHH432 pKa = 4.24VHH434 pKa = 6.41DD435 pKa = 4.91RR436 pKa = 11.84KK437 pKa = 8.93GTYY440 pKa = 9.11IGGYY444 pKa = 7.88YY445 pKa = 9.76QDD447 pKa = 4.18FAFSSVTQSSEE458 pKa = 4.03TGSTPLGTKK467 pKa = 10.01AGQGVSSGSGSIGHH481 pKa = 6.42FHH483 pKa = 6.46TPDD486 pKa = 3.57FGWIQIYY493 pKa = 9.29MSIVPDD499 pKa = 3.55VYY501 pKa = 9.23YY502 pKa = 9.76TQGIPRR508 pKa = 11.84MFSKK512 pKa = 10.66KK513 pKa = 10.39SNLEE517 pKa = 3.6MYY519 pKa = 10.31FPIFNNLPAQAILNKK534 pKa = 9.43EE535 pKa = 4.36LYY537 pKa = 10.54VSGDD541 pKa = 3.27SDD543 pKa = 3.49VDD545 pKa = 3.43NDD547 pKa = 3.39VFAYY551 pKa = 8.88EE552 pKa = 4.61DD553 pKa = 3.91RR554 pKa = 11.84YY555 pKa = 11.09AEE557 pKa = 4.07YY558 pKa = 10.47KK559 pKa = 10.12SRR561 pKa = 11.84HH562 pKa = 4.83NRR564 pKa = 11.84VSGLMALPVDD574 pKa = 3.79KK575 pKa = 10.91APYY578 pKa = 8.49DD579 pKa = 3.68TARR582 pKa = 11.84VIARR586 pKa = 11.84RR587 pKa = 11.84FEE589 pKa = 4.53SVPSLNSLFVTMVPEE604 pKa = 4.19NVDD607 pKa = 3.24MSPFSVTDD615 pKa = 3.72EE616 pKa = 4.36PPFDD620 pKa = 3.28ISVGINCRR628 pKa = 11.84RR629 pKa = 11.84VFPAPYY635 pKa = 8.62TAIEE639 pKa = 4.68GSLSSPALVRR649 pKa = 11.84GG650 pKa = 3.73
MM1 pKa = 7.68ANRR4 pKa = 11.84YY5 pKa = 9.16DD6 pKa = 4.22RR7 pKa = 11.84LDD9 pKa = 3.22NTAPIEE15 pKa = 4.21TALKK19 pKa = 9.98ADD21 pKa = 3.76SVSVGRR27 pKa = 11.84SAFDD31 pKa = 3.08WSCIHH36 pKa = 5.9NTNGIIGSLIPIDD49 pKa = 4.31CFDD52 pKa = 3.72VVPNEE57 pKa = 4.66DD58 pKa = 4.0LDD60 pKa = 3.74ISIASLIEE68 pKa = 3.79FRR70 pKa = 11.84NPTTRR75 pKa = 11.84QLFNGMRR82 pKa = 11.84VFVHH86 pKa = 7.01AYY88 pKa = 8.93YY89 pKa = 10.9NRR91 pKa = 11.84LTDD94 pKa = 3.22LWEE97 pKa = 4.52GARR100 pKa = 11.84NWLDD104 pKa = 3.12NGRR107 pKa = 11.84TGKK110 pKa = 10.75LSLTRR115 pKa = 11.84PNLVYY120 pKa = 10.41TGFYY124 pKa = 9.62KK125 pKa = 10.77APSNVTYY132 pKa = 10.47RR133 pKa = 11.84GNACTPMSLLDD144 pKa = 3.96YY145 pKa = 10.76LGLPAQRR152 pKa = 11.84MQDD155 pKa = 3.38TLKK158 pKa = 10.2TSDD161 pKa = 3.47GSVTASPLRR170 pKa = 11.84SFQCAYY176 pKa = 9.4WSSNNDD182 pKa = 2.99FTASEE187 pKa = 4.31LSYY190 pKa = 11.49APDD193 pKa = 4.54FFPADD198 pKa = 3.17CCMAYY203 pKa = 9.78QRR205 pKa = 11.84NWRR208 pKa = 11.84DD209 pKa = 3.53FYY211 pKa = 11.56CNKK214 pKa = 10.14NLLQNNPYY222 pKa = 9.12WFPEE226 pKa = 4.06NEE228 pKa = 3.75EE229 pKa = 4.26HH230 pKa = 7.71FILSYY235 pKa = 10.3SCSDD239 pKa = 3.55AVCVNYY245 pKa = 10.65GNEE248 pKa = 4.07NLSSVSANALNNIVGNIPTVIADD271 pKa = 3.46YY272 pKa = 11.19SVTPEE277 pKa = 4.09PNNPITNANDD287 pKa = 3.37YY288 pKa = 10.17PGYY291 pKa = 10.23IPTYY295 pKa = 10.12APNLSGLKK303 pKa = 9.72FRR305 pKa = 11.84QFRR308 pKa = 11.84GDD310 pKa = 3.22RR311 pKa = 11.84FTTALPFPDD320 pKa = 5.8LIRR323 pKa = 11.84GDD325 pKa = 3.89IPILEE330 pKa = 4.24YY331 pKa = 10.71TSDD334 pKa = 3.45NVASVTFTRR343 pKa = 11.84SDD345 pKa = 3.64DD346 pKa = 3.94DD347 pKa = 4.27SVEE350 pKa = 4.28LDD352 pKa = 3.18LHH354 pKa = 6.43NNYY357 pKa = 10.34SSTIGGTYY365 pKa = 10.12VNTYY369 pKa = 10.37LKK371 pKa = 10.77DD372 pKa = 3.58STVEE376 pKa = 3.78NTDD379 pKa = 3.53PFNVTATVDD388 pKa = 3.88LSSLGITMSDD398 pKa = 2.92IYY400 pKa = 10.96TLEE403 pKa = 3.83TLTAFRR409 pKa = 11.84RR410 pKa = 11.84RR411 pKa = 11.84MGMTNGDD418 pKa = 4.04YY419 pKa = 11.51NEE421 pKa = 4.62MIKK424 pKa = 10.49AQYY427 pKa = 9.32GVNPHH432 pKa = 4.24VHH434 pKa = 6.41DD435 pKa = 4.91RR436 pKa = 11.84KK437 pKa = 8.93GTYY440 pKa = 9.11IGGYY444 pKa = 7.88YY445 pKa = 9.76QDD447 pKa = 4.18FAFSSVTQSSEE458 pKa = 4.03TGSTPLGTKK467 pKa = 10.01AGQGVSSGSGSIGHH481 pKa = 6.42FHH483 pKa = 6.46TPDD486 pKa = 3.57FGWIQIYY493 pKa = 9.29MSIVPDD499 pKa = 3.55VYY501 pKa = 9.23YY502 pKa = 9.76TQGIPRR508 pKa = 11.84MFSKK512 pKa = 10.66KK513 pKa = 10.39SNLEE517 pKa = 3.6MYY519 pKa = 10.31FPIFNNLPAQAILNKK534 pKa = 9.43EE535 pKa = 4.36LYY537 pKa = 10.54VSGDD541 pKa = 3.27SDD543 pKa = 3.49VDD545 pKa = 3.43NDD547 pKa = 3.39VFAYY551 pKa = 8.88EE552 pKa = 4.61DD553 pKa = 3.91RR554 pKa = 11.84YY555 pKa = 11.09AEE557 pKa = 4.07YY558 pKa = 10.47KK559 pKa = 10.12SRR561 pKa = 11.84HH562 pKa = 4.83NRR564 pKa = 11.84VSGLMALPVDD574 pKa = 3.79KK575 pKa = 10.91APYY578 pKa = 8.49DD579 pKa = 3.68TARR582 pKa = 11.84VIARR586 pKa = 11.84RR587 pKa = 11.84FEE589 pKa = 4.53SVPSLNSLFVTMVPEE604 pKa = 4.19NVDD607 pKa = 3.24MSPFSVTDD615 pKa = 3.72EE616 pKa = 4.36PPFDD620 pKa = 3.28ISVGINCRR628 pKa = 11.84RR629 pKa = 11.84VFPAPYY635 pKa = 8.62TAIEE639 pKa = 4.68GSLSSPALVRR649 pKa = 11.84GG650 pKa = 3.73
Molecular weight: 72.54 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z5CJ66|A0A2Z5CJ66_9VIRU Replication initiation protein OS=Alces alces faeces associated microvirus MP3 6497 OX=2219139 PE=4 SV=1
MM1 pKa = 7.28EE2 pKa = 6.12RR3 pKa = 11.84YY4 pKa = 9.99FNMACARR11 pKa = 11.84PLTILPKK18 pKa = 9.95TRR20 pKa = 11.84FNGDD24 pKa = 3.42YY25 pKa = 9.92IYY27 pKa = 10.59TRR29 pKa = 11.84EE30 pKa = 3.89FTVPCGYY37 pKa = 10.41CLNCRR42 pKa = 11.84RR43 pKa = 11.84DD44 pKa = 3.63YY45 pKa = 11.13QNYY48 pKa = 9.75IIDD51 pKa = 3.82RR52 pKa = 11.84ANYY55 pKa = 7.63EE56 pKa = 3.88YY57 pKa = 10.69CKK59 pKa = 10.28RR60 pKa = 11.84LSASFVTVTYY70 pKa = 11.14DD71 pKa = 3.92DD72 pKa = 3.07IHH74 pKa = 8.39LIEE77 pKa = 4.84HH78 pKa = 6.34CAVRR82 pKa = 11.84DD83 pKa = 3.73YY84 pKa = 11.45DD85 pKa = 3.64GSLIFDD91 pKa = 3.59TDD93 pKa = 3.26SRR95 pKa = 11.84GEE97 pKa = 4.13KK98 pKa = 9.58IVRR101 pKa = 11.84TTLNYY106 pKa = 10.21SDD108 pKa = 3.73FTRR111 pKa = 11.84FVDD114 pKa = 4.39NIRR117 pKa = 11.84HH118 pKa = 5.54YY119 pKa = 10.48IVNHH123 pKa = 5.25SEE125 pKa = 3.86INNILCQPDD134 pKa = 4.37FSYY137 pKa = 10.7LYY139 pKa = 9.97CGEE142 pKa = 4.5YY143 pKa = 10.33GDD145 pKa = 4.57QFGRR149 pKa = 11.84CHH151 pKa = 5.98AHH153 pKa = 6.28FLFFGLDD160 pKa = 3.18FAYY163 pKa = 10.28CKK165 pKa = 10.43KK166 pKa = 10.56IFMDD170 pKa = 2.87KK171 pKa = 10.1WKK173 pKa = 10.6FGFVDD178 pKa = 3.79VLPVLDD184 pKa = 3.66GGIRR188 pKa = 11.84YY189 pKa = 8.2VCKK192 pKa = 10.91YY193 pKa = 8.28MDD195 pKa = 3.58KK196 pKa = 10.85FEE198 pKa = 5.52KK199 pKa = 10.65GFQAEE204 pKa = 4.27LKK206 pKa = 10.54YY207 pKa = 10.79DD208 pKa = 4.17FKK210 pKa = 11.63GLARR214 pKa = 11.84PKK216 pKa = 10.68LRR218 pKa = 11.84MSKK221 pKa = 10.65GFGQGLLWNNAKK233 pKa = 10.35KK234 pKa = 10.33IIEE237 pKa = 4.11NDD239 pKa = 2.95YY240 pKa = 10.8CYY242 pKa = 10.62EE243 pKa = 4.03SRR245 pKa = 11.84HH246 pKa = 5.24KK247 pKa = 9.98QLRR250 pKa = 11.84PISAYY255 pKa = 9.07WKK257 pKa = 10.06LLLTGGVPSRR267 pKa = 11.84DD268 pKa = 3.41VTKK271 pKa = 10.84KK272 pKa = 10.74DD273 pKa = 3.04VWSKK277 pKa = 10.98KK278 pKa = 10.3DD279 pKa = 3.28SYY281 pKa = 10.52LQKK284 pKa = 10.64KK285 pKa = 8.2SASVVRR291 pKa = 11.84SLHH294 pKa = 5.5EE295 pKa = 4.28LNITRR300 pKa = 11.84DD301 pKa = 2.8NGYY304 pKa = 9.25PVDD307 pKa = 3.95SDD309 pKa = 3.58MFQSAFKK316 pKa = 10.36LRR318 pKa = 11.84CARR321 pKa = 11.84IRR323 pKa = 11.84EE324 pKa = 4.21KK325 pKa = 10.89NIEE328 pKa = 4.19VQLHH332 pKa = 6.0NNLTPIPLNHH342 pKa = 6.39IPARR346 pKa = 11.84PKK348 pKa = 10.44FGSITYY354 pKa = 9.87DD355 pKa = 3.22YY356 pKa = 11.07NKK358 pKa = 10.37LKK360 pKa = 10.54KK361 pKa = 10.54CPTRR365 pKa = 11.84TLRR368 pKa = 11.84ALNYY372 pKa = 10.91AFMEE376 pKa = 4.74DD377 pKa = 4.61LFSSLPKK384 pKa = 9.83EE385 pKa = 4.42YY386 pKa = 10.25YY387 pKa = 9.67FHH389 pKa = 8.07SEE391 pKa = 3.87VSS393 pKa = 3.24
MM1 pKa = 7.28EE2 pKa = 6.12RR3 pKa = 11.84YY4 pKa = 9.99FNMACARR11 pKa = 11.84PLTILPKK18 pKa = 9.95TRR20 pKa = 11.84FNGDD24 pKa = 3.42YY25 pKa = 9.92IYY27 pKa = 10.59TRR29 pKa = 11.84EE30 pKa = 3.89FTVPCGYY37 pKa = 10.41CLNCRR42 pKa = 11.84RR43 pKa = 11.84DD44 pKa = 3.63YY45 pKa = 11.13QNYY48 pKa = 9.75IIDD51 pKa = 3.82RR52 pKa = 11.84ANYY55 pKa = 7.63EE56 pKa = 3.88YY57 pKa = 10.69CKK59 pKa = 10.28RR60 pKa = 11.84LSASFVTVTYY70 pKa = 11.14DD71 pKa = 3.92DD72 pKa = 3.07IHH74 pKa = 8.39LIEE77 pKa = 4.84HH78 pKa = 6.34CAVRR82 pKa = 11.84DD83 pKa = 3.73YY84 pKa = 11.45DD85 pKa = 3.64GSLIFDD91 pKa = 3.59TDD93 pKa = 3.26SRR95 pKa = 11.84GEE97 pKa = 4.13KK98 pKa = 9.58IVRR101 pKa = 11.84TTLNYY106 pKa = 10.21SDD108 pKa = 3.73FTRR111 pKa = 11.84FVDD114 pKa = 4.39NIRR117 pKa = 11.84HH118 pKa = 5.54YY119 pKa = 10.48IVNHH123 pKa = 5.25SEE125 pKa = 3.86INNILCQPDD134 pKa = 4.37FSYY137 pKa = 10.7LYY139 pKa = 9.97CGEE142 pKa = 4.5YY143 pKa = 10.33GDD145 pKa = 4.57QFGRR149 pKa = 11.84CHH151 pKa = 5.98AHH153 pKa = 6.28FLFFGLDD160 pKa = 3.18FAYY163 pKa = 10.28CKK165 pKa = 10.43KK166 pKa = 10.56IFMDD170 pKa = 2.87KK171 pKa = 10.1WKK173 pKa = 10.6FGFVDD178 pKa = 3.79VLPVLDD184 pKa = 3.66GGIRR188 pKa = 11.84YY189 pKa = 8.2VCKK192 pKa = 10.91YY193 pKa = 8.28MDD195 pKa = 3.58KK196 pKa = 10.85FEE198 pKa = 5.52KK199 pKa = 10.65GFQAEE204 pKa = 4.27LKK206 pKa = 10.54YY207 pKa = 10.79DD208 pKa = 4.17FKK210 pKa = 11.63GLARR214 pKa = 11.84PKK216 pKa = 10.68LRR218 pKa = 11.84MSKK221 pKa = 10.65GFGQGLLWNNAKK233 pKa = 10.35KK234 pKa = 10.33IIEE237 pKa = 4.11NDD239 pKa = 2.95YY240 pKa = 10.8CYY242 pKa = 10.62EE243 pKa = 4.03SRR245 pKa = 11.84HH246 pKa = 5.24KK247 pKa = 9.98QLRR250 pKa = 11.84PISAYY255 pKa = 9.07WKK257 pKa = 10.06LLLTGGVPSRR267 pKa = 11.84DD268 pKa = 3.41VTKK271 pKa = 10.84KK272 pKa = 10.74DD273 pKa = 3.04VWSKK277 pKa = 10.98KK278 pKa = 10.3DD279 pKa = 3.28SYY281 pKa = 10.52LQKK284 pKa = 10.64KK285 pKa = 8.2SASVVRR291 pKa = 11.84SLHH294 pKa = 5.5EE295 pKa = 4.28LNITRR300 pKa = 11.84DD301 pKa = 2.8NGYY304 pKa = 9.25PVDD307 pKa = 3.95SDD309 pKa = 3.58MFQSAFKK316 pKa = 10.36LRR318 pKa = 11.84CARR321 pKa = 11.84IRR323 pKa = 11.84EE324 pKa = 4.21KK325 pKa = 10.89NIEE328 pKa = 4.19VQLHH332 pKa = 6.0NNLTPIPLNHH342 pKa = 6.39IPARR346 pKa = 11.84PKK348 pKa = 10.44FGSITYY354 pKa = 9.87DD355 pKa = 3.22YY356 pKa = 11.07NKK358 pKa = 10.37LKK360 pKa = 10.54KK361 pKa = 10.54CPTRR365 pKa = 11.84TLRR368 pKa = 11.84ALNYY372 pKa = 10.91AFMEE376 pKa = 4.74DD377 pKa = 4.61LFSSLPKK384 pKa = 9.83EE385 pKa = 4.42YY386 pKa = 10.25YY387 pKa = 9.67FHH389 pKa = 8.07SEE391 pKa = 3.87VSS393 pKa = 3.24
Molecular weight: 46.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1363 |
320 |
650 |
454.3 |
51.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.236 ± 0.911 | 1.761 ± 0.785 |
7.117 ± 0.091 | 4.035 ± 0.194 |
5.062 ± 0.779 | 6.163 ± 0.538 |
1.761 ± 0.376 | 5.723 ± 0.338 |
4.916 ± 1.447 | 7.85 ± 0.349 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.861 ± 0.888 | 7.483 ± 0.891 |
4.549 ± 1.135 | 3.302 ± 1.287 |
5.356 ± 0.814 | 8.291 ± 0.869 |
5.943 ± 0.722 | 4.842 ± 1.247 |
1.174 ± 0.14 | 5.576 ± 1.051 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
