
Besnoitia besnoiti
Taxonomy: cellular organisms; Eukaryota; Sar; Alveolata; Apicomplexa; Conoidasida; Coccidia; Eucoccidiorida; Eimeriorina; Sarcocystidae; Besnoitia
Average proteome isoelectric point is 6.81
Get precalculated fractions of proteins
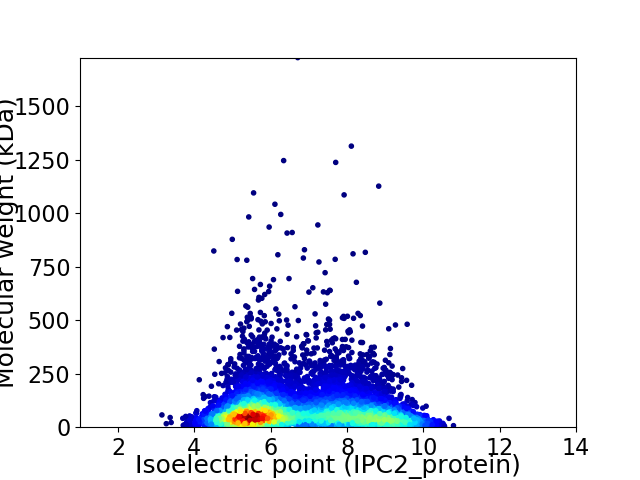
Virtual 2D-PAGE plot for 8036 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2A9MMI2|A0A2A9MMI2_9APIC Uncharacterized protein OS=Besnoitia besnoiti OX=94643 GN=BESB_008960 PE=4 SV=1
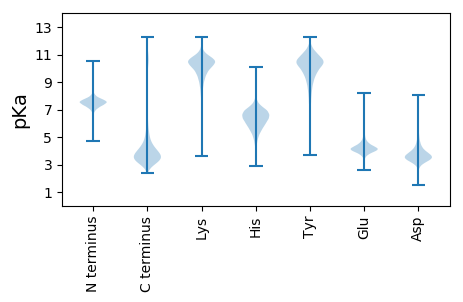
MM1 pKa = 7.27EE2 pKa = 5.58RR3 pKa = 11.84SNPRR7 pKa = 11.84LEE9 pKa = 4.44RR10 pKa = 11.84SNPDD14 pKa = 2.66WRR16 pKa = 11.84DD17 pKa = 3.21PAPVWRR23 pKa = 11.84SRR25 pKa = 11.84APDD28 pKa = 3.13WRR30 pKa = 11.84SRR32 pKa = 11.84APDD35 pKa = 3.15WRR37 pKa = 11.84NPAPHH42 pKa = 6.69WRR44 pKa = 11.84DD45 pKa = 3.34GTPHH49 pKa = 5.86WRR51 pKa = 11.84NRR53 pKa = 11.84APTGEE58 pKa = 4.09VEE60 pKa = 4.17PPYY63 pKa = 11.23GEE65 pKa = 3.96IQPPTGEE72 pKa = 4.05VEE74 pKa = 4.0PPYY77 pKa = 11.21GEE79 pKa = 4.44VEE81 pKa = 4.2PPTGEE86 pKa = 3.58IEE88 pKa = 4.05PPYY91 pKa = 10.79EE92 pKa = 4.17EE93 pKa = 5.01VEE95 pKa = 4.11PPYY98 pKa = 11.3GEE100 pKa = 4.44VEE102 pKa = 4.21PPTGEE107 pKa = 3.64IQPPYY112 pKa = 11.03GEE114 pKa = 4.47IEE116 pKa = 3.96PPYY119 pKa = 10.99GEE121 pKa = 4.52VEE123 pKa = 4.21PPTGEE128 pKa = 3.67IQPPNGEE135 pKa = 4.18IEE137 pKa = 4.28PPTGEE142 pKa = 3.98VEE144 pKa = 4.07PPYY147 pKa = 11.23GEE149 pKa = 3.96IQPPTGEE156 pKa = 4.05IQPPTGEE163 pKa = 3.82IQPPYY168 pKa = 10.95GEE170 pKa = 4.55VEE172 pKa = 4.13PPTGEE177 pKa = 4.08VEE179 pKa = 4.22PPTGEE184 pKa = 3.72IQLPTGEE191 pKa = 4.6MEE193 pKa = 4.53PPTGEE198 pKa = 3.88IEE200 pKa = 4.25PPTGEE205 pKa = 3.98VEE207 pKa = 4.05PPYY210 pKa = 11.2GEE212 pKa = 4.49VEE214 pKa = 4.16PTTGEE219 pKa = 3.67IQPPYY224 pKa = 11.03GEE226 pKa = 4.47IEE228 pKa = 3.96PPYY231 pKa = 10.99GEE233 pKa = 4.52VEE235 pKa = 4.21PPTGEE240 pKa = 3.64IQPPYY245 pKa = 11.03GEE247 pKa = 4.47IEE249 pKa = 3.96PPYY252 pKa = 10.99GEE254 pKa = 4.52VEE256 pKa = 4.21PPTGEE261 pKa = 3.64IQPPYY266 pKa = 10.95GEE268 pKa = 4.55VEE270 pKa = 4.13PPTGEE275 pKa = 3.93VEE277 pKa = 4.05PPYY280 pKa = 11.23GEE282 pKa = 4.37VEE284 pKa = 4.02PPYY287 pKa = 11.22GEE289 pKa = 3.96IQPPTGEE296 pKa = 3.91IQTPDD301 pKa = 3.07WRR303 pKa = 11.84DD304 pKa = 3.26PAPP307 pKa = 3.81
MM1 pKa = 7.27EE2 pKa = 5.58RR3 pKa = 11.84SNPRR7 pKa = 11.84LEE9 pKa = 4.44RR10 pKa = 11.84SNPDD14 pKa = 2.66WRR16 pKa = 11.84DD17 pKa = 3.21PAPVWRR23 pKa = 11.84SRR25 pKa = 11.84APDD28 pKa = 3.13WRR30 pKa = 11.84SRR32 pKa = 11.84APDD35 pKa = 3.15WRR37 pKa = 11.84NPAPHH42 pKa = 6.69WRR44 pKa = 11.84DD45 pKa = 3.34GTPHH49 pKa = 5.86WRR51 pKa = 11.84NRR53 pKa = 11.84APTGEE58 pKa = 4.09VEE60 pKa = 4.17PPYY63 pKa = 11.23GEE65 pKa = 3.96IQPPTGEE72 pKa = 4.05VEE74 pKa = 4.0PPYY77 pKa = 11.21GEE79 pKa = 4.44VEE81 pKa = 4.2PPTGEE86 pKa = 3.58IEE88 pKa = 4.05PPYY91 pKa = 10.79EE92 pKa = 4.17EE93 pKa = 5.01VEE95 pKa = 4.11PPYY98 pKa = 11.3GEE100 pKa = 4.44VEE102 pKa = 4.21PPTGEE107 pKa = 3.64IQPPYY112 pKa = 11.03GEE114 pKa = 4.47IEE116 pKa = 3.96PPYY119 pKa = 10.99GEE121 pKa = 4.52VEE123 pKa = 4.21PPTGEE128 pKa = 3.67IQPPNGEE135 pKa = 4.18IEE137 pKa = 4.28PPTGEE142 pKa = 3.98VEE144 pKa = 4.07PPYY147 pKa = 11.23GEE149 pKa = 3.96IQPPTGEE156 pKa = 4.05IQPPTGEE163 pKa = 3.82IQPPYY168 pKa = 10.95GEE170 pKa = 4.55VEE172 pKa = 4.13PPTGEE177 pKa = 4.08VEE179 pKa = 4.22PPTGEE184 pKa = 3.72IQLPTGEE191 pKa = 4.6MEE193 pKa = 4.53PPTGEE198 pKa = 3.88IEE200 pKa = 4.25PPTGEE205 pKa = 3.98VEE207 pKa = 4.05PPYY210 pKa = 11.2GEE212 pKa = 4.49VEE214 pKa = 4.16PTTGEE219 pKa = 3.67IQPPYY224 pKa = 11.03GEE226 pKa = 4.47IEE228 pKa = 3.96PPYY231 pKa = 10.99GEE233 pKa = 4.52VEE235 pKa = 4.21PPTGEE240 pKa = 3.64IQPPYY245 pKa = 11.03GEE247 pKa = 4.47IEE249 pKa = 3.96PPYY252 pKa = 10.99GEE254 pKa = 4.52VEE256 pKa = 4.21PPTGEE261 pKa = 3.64IQPPYY266 pKa = 10.95GEE268 pKa = 4.55VEE270 pKa = 4.13PPTGEE275 pKa = 3.93VEE277 pKa = 4.05PPYY280 pKa = 11.23GEE282 pKa = 4.37VEE284 pKa = 4.02PPYY287 pKa = 11.22GEE289 pKa = 3.96IQPPTGEE296 pKa = 3.91IQTPDD301 pKa = 3.07WRR303 pKa = 11.84DD304 pKa = 3.26PAPP307 pKa = 3.81
Molecular weight: 33.8 kDa
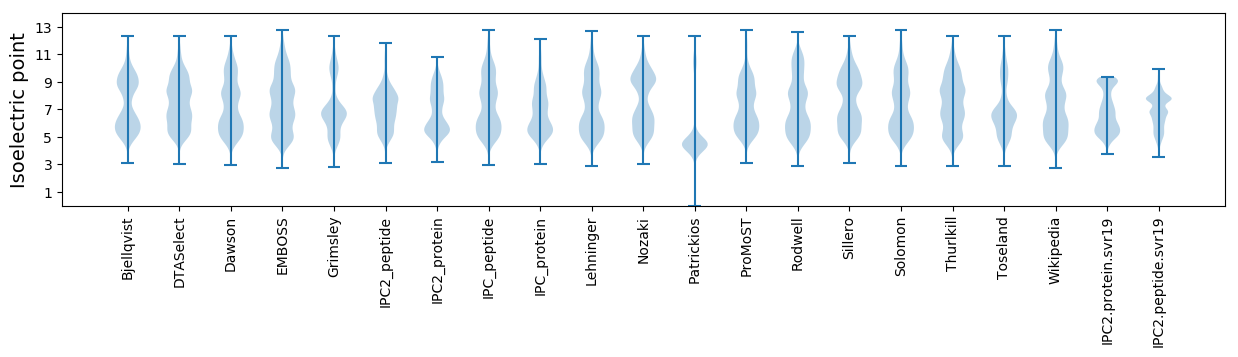
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2A9MI01|A0A2A9MI01_9APIC Ubiquitin carboxyl-terminal hydrolase OS=Besnoitia besnoiti OX=94643 GN=BESB_055320 PE=4 SV=1
MM1 pKa = 6.41VAARR5 pKa = 11.84AEE7 pKa = 4.11EE8 pKa = 4.03RR9 pKa = 11.84RR10 pKa = 11.84VCSPPSSAEE19 pKa = 4.0SSSSLSAFSEE29 pKa = 4.28GRR31 pKa = 11.84GRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84PRR37 pKa = 11.84EE38 pKa = 3.88KK39 pKa = 10.91APATAALDD47 pKa = 3.71RR48 pKa = 11.84FGLFSQTQSPWTTLAAPAFCTTGSVSRR75 pKa = 11.84YY76 pKa = 6.65EE77 pKa = 4.03QLVAAIKK84 pKa = 9.77EE85 pKa = 4.04KK86 pKa = 10.99HH87 pKa = 6.23KK88 pKa = 10.86GRR90 pKa = 11.84KK91 pKa = 6.41TKK93 pKa = 9.51EE94 pKa = 3.43KK95 pKa = 10.43RR96 pKa = 11.84RR97 pKa = 11.84ISEE100 pKa = 4.0GLLLFLAAVTHH111 pKa = 6.72GGTAALSRR119 pKa = 11.84LLLAPLDD126 pKa = 3.55QEE128 pKa = 4.6KK129 pKa = 10.39IIRR132 pKa = 11.84QLSLARR138 pKa = 11.84LPSLSPSSSPWSPSPQGSAVCASPFLVSEE167 pKa = 4.39SASPSSSAPVRR178 pKa = 11.84ARR180 pKa = 11.84RR181 pKa = 11.84PQSSSALSSGPPSSASPPSSSVSFSAARR209 pKa = 11.84ARR211 pKa = 11.84ASAASSAPNSSVKK224 pKa = 10.3PPSAPAGASIRR235 pKa = 11.84PSPSANSAAAFPTSGLPSASSLPPSRR261 pKa = 11.84WLTVSSFWLGCSAPICAAIGGSWIRR286 pKa = 11.84LALYY290 pKa = 9.97QRR292 pKa = 11.84TRR294 pKa = 11.84LFFTDD299 pKa = 4.05GEE301 pKa = 4.5EE302 pKa = 4.43TYY304 pKa = 11.58SNAEE308 pKa = 3.6RR309 pKa = 11.84FFRR312 pKa = 11.84NMGCICFSAFVALAVCYY329 pKa = 9.47PLDD332 pKa = 3.7VAHH335 pKa = 6.72TAMCVIAASARR346 pKa = 11.84QSGRR350 pKa = 11.84VISSLDD356 pKa = 3.24AFSAFSATSAPQGARR371 pKa = 11.84RR372 pKa = 11.84EE373 pKa = 4.13ASVSFLSRR381 pKa = 11.84RR382 pKa = 11.84FASAKK387 pKa = 9.78KK388 pKa = 10.24RR389 pKa = 11.84LGAQLTGTVTTGSARR404 pKa = 11.84AEE406 pKa = 4.39TVDD409 pKa = 3.32GEE411 pKa = 3.99ARR413 pKa = 11.84QRR415 pKa = 11.84HH416 pKa = 4.91ASARR420 pKa = 11.84AVSPSHH426 pKa = 6.36IVWRR430 pKa = 11.84LYY432 pKa = 10.25KK433 pKa = 10.32QGGVRR438 pKa = 11.84ALYY441 pKa = 10.58CGFGLCALTLVPFTLASAFLQLKK464 pKa = 8.29FQSLLLSLQQQEE476 pKa = 4.73GAVAAHH482 pKa = 6.98WPRR485 pKa = 11.84LFASRR490 pKa = 11.84NGDD493 pKa = 3.62LPLACDD499 pKa = 3.52APKK502 pKa = 11.01GDD504 pKa = 3.58GRR506 pKa = 11.84GADD509 pKa = 3.84EE510 pKa = 4.58EE511 pKa = 4.74EE512 pKa = 3.98EE513 pKa = 4.22DD514 pKa = 4.02ARR516 pKa = 11.84GLSRR520 pKa = 11.84EE521 pKa = 4.14KK522 pKa = 10.58EE523 pKa = 3.95RR524 pKa = 11.84SPVADD529 pKa = 3.85EE530 pKa = 3.91TATAGDD536 pKa = 3.22QGAAPRR542 pKa = 11.84RR543 pKa = 11.84RR544 pKa = 11.84KK545 pKa = 8.52TGGKK549 pKa = 8.22RR550 pKa = 11.84QAVRR554 pKa = 11.84GDD556 pKa = 3.22ADD558 pKa = 3.47EE559 pKa = 4.23TNRR562 pKa = 11.84AGEE565 pKa = 4.25SASEE569 pKa = 4.36DD570 pKa = 3.49GRR572 pKa = 11.84EE573 pKa = 3.79TTYY576 pKa = 10.73PRR578 pKa = 11.84GRR580 pKa = 11.84GGSAFRR586 pKa = 11.84QARR589 pKa = 11.84GEE591 pKa = 4.16IAHH594 pKa = 6.5SPQTDD599 pKa = 2.98SFEE602 pKa = 4.58RR603 pKa = 11.84KK604 pKa = 7.72RR605 pKa = 11.84HH606 pKa = 4.78EE607 pKa = 4.17EE608 pKa = 3.89RR609 pKa = 11.84EE610 pKa = 4.14EE611 pKa = 3.99TLGSSRR617 pKa = 11.84WKK619 pKa = 10.47AAATAAFAAGLVAQLATYY637 pKa = 9.53PLDD640 pKa = 3.98SIRR643 pKa = 11.84RR644 pKa = 11.84RR645 pKa = 11.84QQFATISSLLFPHH658 pKa = 6.84GVAAPSRR665 pKa = 11.84VQPPASPMSVHH676 pKa = 6.47PPKK679 pKa = 10.06PVSGFRR685 pKa = 11.84RR686 pKa = 11.84FFVVPSSRR694 pKa = 11.84AWFSKK699 pKa = 9.33PHH701 pKa = 6.12GGLFRR706 pKa = 11.84GAGVLLVRR714 pKa = 11.84SVPEE718 pKa = 3.51CAIAVSVYY726 pKa = 10.42SFLMSKK732 pKa = 9.27MPSLAYY738 pKa = 10.08HH739 pKa = 6.93
MM1 pKa = 6.41VAARR5 pKa = 11.84AEE7 pKa = 4.11EE8 pKa = 4.03RR9 pKa = 11.84RR10 pKa = 11.84VCSPPSSAEE19 pKa = 4.0SSSSLSAFSEE29 pKa = 4.28GRR31 pKa = 11.84GRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84PRR37 pKa = 11.84EE38 pKa = 3.88KK39 pKa = 10.91APATAALDD47 pKa = 3.71RR48 pKa = 11.84FGLFSQTQSPWTTLAAPAFCTTGSVSRR75 pKa = 11.84YY76 pKa = 6.65EE77 pKa = 4.03QLVAAIKK84 pKa = 9.77EE85 pKa = 4.04KK86 pKa = 10.99HH87 pKa = 6.23KK88 pKa = 10.86GRR90 pKa = 11.84KK91 pKa = 6.41TKK93 pKa = 9.51EE94 pKa = 3.43KK95 pKa = 10.43RR96 pKa = 11.84RR97 pKa = 11.84ISEE100 pKa = 4.0GLLLFLAAVTHH111 pKa = 6.72GGTAALSRR119 pKa = 11.84LLLAPLDD126 pKa = 3.55QEE128 pKa = 4.6KK129 pKa = 10.39IIRR132 pKa = 11.84QLSLARR138 pKa = 11.84LPSLSPSSSPWSPSPQGSAVCASPFLVSEE167 pKa = 4.39SASPSSSAPVRR178 pKa = 11.84ARR180 pKa = 11.84RR181 pKa = 11.84PQSSSALSSGPPSSASPPSSSVSFSAARR209 pKa = 11.84ARR211 pKa = 11.84ASAASSAPNSSVKK224 pKa = 10.3PPSAPAGASIRR235 pKa = 11.84PSPSANSAAAFPTSGLPSASSLPPSRR261 pKa = 11.84WLTVSSFWLGCSAPICAAIGGSWIRR286 pKa = 11.84LALYY290 pKa = 9.97QRR292 pKa = 11.84TRR294 pKa = 11.84LFFTDD299 pKa = 4.05GEE301 pKa = 4.5EE302 pKa = 4.43TYY304 pKa = 11.58SNAEE308 pKa = 3.6RR309 pKa = 11.84FFRR312 pKa = 11.84NMGCICFSAFVALAVCYY329 pKa = 9.47PLDD332 pKa = 3.7VAHH335 pKa = 6.72TAMCVIAASARR346 pKa = 11.84QSGRR350 pKa = 11.84VISSLDD356 pKa = 3.24AFSAFSATSAPQGARR371 pKa = 11.84RR372 pKa = 11.84EE373 pKa = 4.13ASVSFLSRR381 pKa = 11.84RR382 pKa = 11.84FASAKK387 pKa = 9.78KK388 pKa = 10.24RR389 pKa = 11.84LGAQLTGTVTTGSARR404 pKa = 11.84AEE406 pKa = 4.39TVDD409 pKa = 3.32GEE411 pKa = 3.99ARR413 pKa = 11.84QRR415 pKa = 11.84HH416 pKa = 4.91ASARR420 pKa = 11.84AVSPSHH426 pKa = 6.36IVWRR430 pKa = 11.84LYY432 pKa = 10.25KK433 pKa = 10.32QGGVRR438 pKa = 11.84ALYY441 pKa = 10.58CGFGLCALTLVPFTLASAFLQLKK464 pKa = 8.29FQSLLLSLQQQEE476 pKa = 4.73GAVAAHH482 pKa = 6.98WPRR485 pKa = 11.84LFASRR490 pKa = 11.84NGDD493 pKa = 3.62LPLACDD499 pKa = 3.52APKK502 pKa = 11.01GDD504 pKa = 3.58GRR506 pKa = 11.84GADD509 pKa = 3.84EE510 pKa = 4.58EE511 pKa = 4.74EE512 pKa = 3.98EE513 pKa = 4.22DD514 pKa = 4.02ARR516 pKa = 11.84GLSRR520 pKa = 11.84EE521 pKa = 4.14KK522 pKa = 10.58EE523 pKa = 3.95RR524 pKa = 11.84SPVADD529 pKa = 3.85EE530 pKa = 3.91TATAGDD536 pKa = 3.22QGAAPRR542 pKa = 11.84RR543 pKa = 11.84RR544 pKa = 11.84KK545 pKa = 8.52TGGKK549 pKa = 8.22RR550 pKa = 11.84QAVRR554 pKa = 11.84GDD556 pKa = 3.22ADD558 pKa = 3.47EE559 pKa = 4.23TNRR562 pKa = 11.84AGEE565 pKa = 4.25SASEE569 pKa = 4.36DD570 pKa = 3.49GRR572 pKa = 11.84EE573 pKa = 3.79TTYY576 pKa = 10.73PRR578 pKa = 11.84GRR580 pKa = 11.84GGSAFRR586 pKa = 11.84QARR589 pKa = 11.84GEE591 pKa = 4.16IAHH594 pKa = 6.5SPQTDD599 pKa = 2.98SFEE602 pKa = 4.58RR603 pKa = 11.84KK604 pKa = 7.72RR605 pKa = 11.84HH606 pKa = 4.78EE607 pKa = 4.17EE608 pKa = 3.89RR609 pKa = 11.84EE610 pKa = 4.14EE611 pKa = 3.99TLGSSRR617 pKa = 11.84WKK619 pKa = 10.47AAATAAFAAGLVAQLATYY637 pKa = 9.53PLDD640 pKa = 3.98SIRR643 pKa = 11.84RR644 pKa = 11.84RR645 pKa = 11.84QQFATISSLLFPHH658 pKa = 6.84GVAAPSRR665 pKa = 11.84VQPPASPMSVHH676 pKa = 6.47PPKK679 pKa = 10.06PVSGFRR685 pKa = 11.84RR686 pKa = 11.84FFVVPSSRR694 pKa = 11.84AWFSKK699 pKa = 9.33PHH701 pKa = 6.12GGLFRR706 pKa = 11.84GAGVLLVRR714 pKa = 11.84SVPEE718 pKa = 3.51CAIAVSVYY726 pKa = 10.42SFLMSKK732 pKa = 9.27MPSLAYY738 pKa = 10.08HH739 pKa = 6.93
Molecular weight: 78.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6780256 |
18 |
16347 |
843.7 |
90.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.892 ± 0.049 | 1.938 ± 0.015 |
4.759 ± 0.017 | 7.202 ± 0.03 |
3.182 ± 0.016 | 8.073 ± 0.028 |
2.089 ± 0.011 | 2.269 ± 0.02 |
3.59 ± 0.029 | 8.985 ± 0.029 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.329 ± 0.01 | 1.978 ± 0.015 |
6.937 ± 0.029 | 3.803 ± 0.017 |
8.257 ± 0.035 | 10.167 ± 0.04 |
4.539 ± 0.016 | 5.514 ± 0.022 |
0.998 ± 0.006 | 1.498 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
