
Methanoculleus bourgensis (strain ATCC 43281 / DSM 3045 / OCM 15 / MS2) (Methanogenium bourgense)
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Methanomicrobia; Methanomicrobiales; Methanomicrobiaceae; Methanoculleus; Methanoculleus bourgensis
Average proteome isoelectric point is 5.96
Get precalculated fractions of proteins
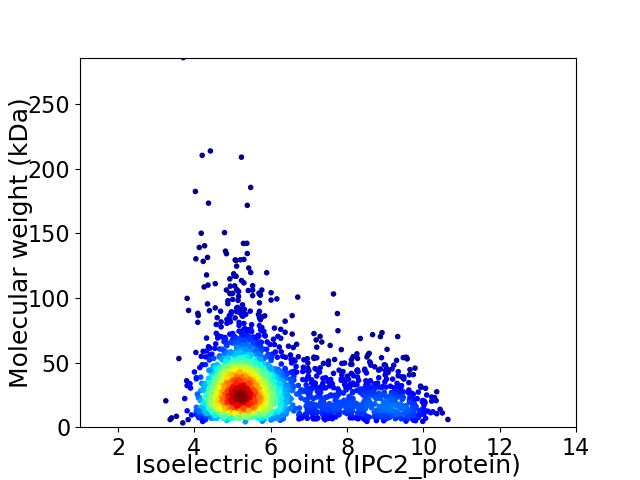
Virtual 2D-PAGE plot for 2575 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I7KDV4|I7KDV4_METBM Hydrogenase 1 maturation protease OS=Methanoculleus bourgensis (strain ATCC 43281 / DSM 3045 / OCM 15 / MS2) OX=1201294 GN=hycI PE=4 SV=2
MM1 pKa = 7.74DD2 pKa = 5.2RR3 pKa = 11.84SRR5 pKa = 11.84VIAGAIVILLAVAVLAVPVAAEE27 pKa = 4.09EE28 pKa = 4.54YY29 pKa = 10.88VFVTKK34 pKa = 10.1WGEE37 pKa = 3.93YY38 pKa = 8.58GTGEE42 pKa = 4.23GFKK45 pKa = 10.93DD46 pKa = 3.63PFDD49 pKa = 3.8IAIDD53 pKa = 3.97TAGHH57 pKa = 6.34IYY59 pKa = 8.73ITEE62 pKa = 3.84TDD64 pKa = 3.62ANHH67 pKa = 6.18ILQRR71 pKa = 11.84ILKK74 pKa = 9.12LDD76 pKa = 3.18SSMNFITKK84 pKa = 8.8WGSYY88 pKa = 7.84GTGNGQFNGPQGIAVNAAGNVYY110 pKa = 10.43VADD113 pKa = 4.42TYY115 pKa = 9.83NHH117 pKa = 7.6RR118 pKa = 11.84IQKK121 pKa = 9.75FDD123 pKa = 3.1SSGNLLTKK131 pKa = 9.37WGSWGSGDD139 pKa = 4.14GQFSYY144 pKa = 10.58PDD146 pKa = 3.6SVAVDD151 pKa = 3.35AAGNVYY157 pKa = 10.42VSDD160 pKa = 3.96TNNGRR165 pKa = 11.84IQKK168 pKa = 9.52FDD170 pKa = 3.43SDD172 pKa = 4.0GTFLGKK178 pKa = 9.32WGSWGSGDD186 pKa = 3.38GQLRR190 pKa = 11.84FSQDD194 pKa = 3.26LVVDD198 pKa = 4.21AAGNIYY204 pKa = 9.32VAEE207 pKa = 4.22YY208 pKa = 10.46GNHH211 pKa = 7.16RR212 pKa = 11.84IQKK215 pKa = 9.55FDD217 pKa = 3.87SNGNFLWKK225 pKa = 10.09KK226 pKa = 9.5GSSGSGDD233 pKa = 3.57GQFLSPYY240 pKa = 9.73GITVDD245 pKa = 3.28AAGNVYY251 pKa = 10.41VADD254 pKa = 3.82TWNHH258 pKa = 7.0RR259 pKa = 11.84IQKK262 pKa = 9.56FDD264 pKa = 3.09SSGNFLTKK272 pKa = 9.65WGSRR276 pKa = 11.84GSGNGQFSEE285 pKa = 4.43PFGVAVDD292 pKa = 3.82SAGNVYY298 pKa = 8.42VTEE301 pKa = 4.42RR302 pKa = 11.84GNDD305 pKa = 3.32RR306 pKa = 11.84VQMFAPASALTVTGIVPASGLNTGSVSITDD336 pKa = 3.72LSGTGFANGATVNLTRR352 pKa = 11.84TGEE355 pKa = 4.26TNIVATDD362 pKa = 3.41VSVLSSTQITCTLDD376 pKa = 3.07LTGAVAGSWNVVVTNPDD393 pKa = 3.13NQFATLTDD401 pKa = 3.69GFTVNAEE408 pKa = 3.46NRR410 pKa = 11.84APVAEE415 pKa = 5.2DD416 pKa = 3.09DD417 pKa = 4.41TYY419 pKa = 9.7TTDD422 pKa = 3.77EE423 pKa = 4.57DD424 pKa = 4.22TLLTISAPGVLANDD438 pKa = 3.76NDD440 pKa = 4.41PDD442 pKa = 4.47GDD444 pKa = 4.02ALTAVLVDD452 pKa = 3.99DD453 pKa = 4.8VSNGTLTLNADD464 pKa = 3.48GSFTYY469 pKa = 10.58LPNTGFSDD477 pKa = 3.2IDD479 pKa = 3.77TFTYY483 pKa = 8.55TANDD487 pKa = 3.9GTQSSNVATVSINVNPASAVLTSIVVTPVQPILEE521 pKa = 4.34IGGTQQFTATGYY533 pKa = 10.12DD534 pKa = 3.35QYY536 pKa = 11.15GTPLPTGEE544 pKa = 4.62IVWSSTNTTVGTIDD558 pKa = 3.17ATGLFTATAEE568 pKa = 4.3GSTTVTATAGEE579 pKa = 4.26VSGTAEE585 pKa = 4.1VTVNPALTVTGIVPASGPNTGNVSITNLSGTGFVDD620 pKa = 4.44GATVNLTRR628 pKa = 11.84TGEE631 pKa = 4.26TNIVATDD638 pKa = 3.41VSVLSSTQITCTLDD652 pKa = 3.24LTGAAAGSWNVVVTNPDD669 pKa = 2.55GRR671 pKa = 11.84YY672 pKa = 10.25DD673 pKa = 3.45ILSDD677 pKa = 3.65GFAVVAASEE686 pKa = 4.23IVTFNDD692 pKa = 3.24PNLEE696 pKa = 4.03ATVRR700 pKa = 11.84GALGKK705 pKa = 9.62PVGDD709 pKa = 3.48ITADD713 pKa = 3.44DD714 pKa = 3.87MATFITLGADD724 pKa = 2.75WRR726 pKa = 11.84GIRR729 pKa = 11.84DD730 pKa = 3.66LSGLEE735 pKa = 3.91YY736 pKa = 10.66AVNLQHH742 pKa = 7.68LYY744 pKa = 10.25LQQNRR749 pKa = 11.84QISDD753 pKa = 4.05LGPLAGLTDD762 pKa = 4.68LQTLDD767 pKa = 3.55LWNNQISDD775 pKa = 4.32LSPLAGLTNLSVLLLGSNQISDD797 pKa = 3.61IGPLAGLTDD806 pKa = 4.23LQRR809 pKa = 11.84LHH811 pKa = 7.56LYY813 pKa = 10.46DD814 pKa = 3.53NQIRR818 pKa = 11.84DD819 pKa = 3.63IGPLAGLTNLWEE831 pKa = 3.94LRR833 pKa = 11.84LYY835 pKa = 10.85NNQIRR840 pKa = 11.84DD841 pKa = 3.57IGPLVANSGLGSGDD855 pKa = 3.48EE856 pKa = 4.86VYY858 pKa = 10.8LQYY861 pKa = 11.34NYY863 pKa = 11.17LDD865 pKa = 4.18LTPGSADD872 pKa = 3.31MNDD875 pKa = 2.83IQTLQSRR882 pKa = 11.84GVYY885 pKa = 8.77VAYY888 pKa = 9.89EE889 pKa = 4.02PQNPVPTYY897 pKa = 10.22TLDD900 pKa = 3.64LAVNPEE906 pKa = 3.93GGGTVTGAGTYY917 pKa = 9.84KK918 pKa = 10.78AGDD921 pKa = 3.91TVPITATPNEE931 pKa = 3.83GWEE934 pKa = 4.21FVNWTNEE941 pKa = 3.44TGATVSSEE949 pKa = 4.17PNFDD953 pKa = 3.5YY954 pKa = 10.84PMPEE958 pKa = 3.72GDD960 pKa = 3.79VALTANFEE968 pKa = 4.47GEE970 pKa = 4.52AITYY974 pKa = 7.91TLGLAVNPAGSGTVTGAGTYY994 pKa = 10.21AAGDD998 pKa = 4.11TVSVTATPNDD1008 pKa = 3.93GYY1010 pKa = 9.63TFVNWTDD1017 pKa = 3.45EE1018 pKa = 4.19TDD1020 pKa = 3.76VTVSTAASFDD1030 pKa = 3.5YY1031 pKa = 10.74TMPDD1035 pKa = 2.86GDD1037 pKa = 3.87VTLTANFAAITYY1049 pKa = 8.43TLDD1052 pKa = 3.49LAVNPEE1058 pKa = 3.99GSGTATGAGTYY1069 pKa = 10.09AAGATVPITATPNEE1083 pKa = 4.08GWEE1086 pKa = 4.05FANWTDD1092 pKa = 3.56GTGATVSSEE1101 pKa = 3.8ATFDD1105 pKa = 3.54YY1106 pKa = 10.86TMPAQEE1112 pKa = 4.32TALTANFEE1120 pKa = 4.16PAPPVLARR1128 pKa = 11.84IEE1130 pKa = 4.1VSPAEE1135 pKa = 4.01ATLEE1139 pKa = 4.14VGATRR1144 pKa = 11.84QFTATGYY1151 pKa = 10.08DD1152 pKa = 3.37QYY1154 pKa = 11.8GNPLSVGEE1162 pKa = 4.53IVWSSTNEE1170 pKa = 3.67AVGTIDD1176 pKa = 3.56ASGMFTALAAGSTEE1190 pKa = 4.47LVAEE1194 pKa = 4.35ANGISGKK1201 pKa = 10.84ANVTVTPAAPVLTSIAVTPEE1221 pKa = 3.85SPALEE1226 pKa = 4.43IGTTQQFTATCYY1238 pKa = 10.15DD1239 pKa = 3.23QYY1241 pKa = 10.97GQTMPDD1247 pKa = 3.31MAVSWSSSNTSVGTIEE1263 pKa = 4.28SNGMFTALTAGSTTVTASAAGVSGQTVATVNPAAPVVTRR1302 pKa = 11.84IAVSPPSVTLDD1313 pKa = 3.29IGDD1316 pKa = 3.81TQAFVATCYY1325 pKa = 10.42DD1326 pKa = 3.23QYY1328 pKa = 10.97EE1329 pKa = 4.3NEE1331 pKa = 4.23MPGTSVSWASDD1342 pKa = 3.09STTVGTIDD1350 pKa = 3.17ATGLFTATAEE1360 pKa = 4.3GSTTVTATAGDD1371 pKa = 3.63VSGTAEE1377 pKa = 4.03VRR1379 pKa = 11.84VNPALTVTGIVPASGPNTGPVSITDD1404 pKa = 3.71LSGTGFADD1412 pKa = 4.16GATVKK1417 pKa = 9.63LTRR1420 pKa = 11.84DD1421 pKa = 3.34GEE1423 pKa = 4.18ADD1425 pKa = 3.15IVATDD1430 pKa = 3.64VAVLSPTQITCTLDD1444 pKa = 3.16LTGAAVGTWNVVVTNPDD1461 pKa = 2.99EE1462 pKa = 4.45QYY1464 pKa = 11.29DD1465 pKa = 3.73ILSDD1469 pKa = 3.81GFAVVAASEE1478 pKa = 4.52VVTFNDD1484 pKa = 3.69PNLEE1488 pKa = 3.85AAVRR1492 pKa = 11.84QEE1494 pKa = 3.65LSKK1497 pKa = 11.07PKK1499 pKa = 10.75GDD1501 pKa = 3.28ITADD1505 pKa = 3.42DD1506 pKa = 4.1MATLTRR1512 pKa = 11.84LSAAGRR1518 pKa = 11.84GIRR1521 pKa = 11.84DD1522 pKa = 3.53LSGLEE1527 pKa = 4.0YY1528 pKa = 10.64AVNLQTLYY1536 pKa = 10.51ISNNQISDD1544 pKa = 4.09LSPLAGLTNLQTLWLQDD1561 pKa = 3.53NQVSDD1566 pKa = 4.43LSPLAGLTNLQRR1578 pKa = 11.84LWLNQNQIRR1587 pKa = 11.84DD1588 pKa = 3.78VSPLAGLTNLRR1599 pKa = 11.84EE1600 pKa = 4.02LLLAVNQISDD1610 pKa = 4.26LSPLAGLTNLGYY1622 pKa = 10.21VQLYY1626 pKa = 9.85RR1627 pKa = 11.84NQISDD1632 pKa = 4.26LSPLAGLTNLGYY1644 pKa = 10.21VQLYY1648 pKa = 9.85RR1649 pKa = 11.84NQISDD1654 pKa = 4.26LSPLAGLTNLYY1665 pKa = 10.49FLDD1668 pKa = 3.21ISYY1671 pKa = 10.43NQISDD1676 pKa = 4.26LSPLASLTNLYY1687 pKa = 10.55FLDD1690 pKa = 3.21ISYY1693 pKa = 10.4NQISDD1698 pKa = 3.68ISPLAGLTRR1707 pKa = 11.84LSRR1710 pKa = 11.84LSLDD1714 pKa = 3.46NNQISDD1720 pKa = 3.8ISPLAGLINLYY1731 pKa = 10.12VLNLNYY1737 pKa = 10.58NQIRR1741 pKa = 11.84DD1742 pKa = 3.76ISPLVANSGLAGDD1755 pKa = 5.2DD1756 pKa = 3.85VYY1758 pKa = 11.61LQYY1761 pKa = 11.36NYY1763 pKa = 11.1LDD1765 pKa = 4.08LTPGSAAMNDD1775 pKa = 3.04IQTLQSRR1782 pKa = 11.84GVYY1785 pKa = 9.2VVYY1788 pKa = 10.25EE1789 pKa = 4.13PQHH1792 pKa = 6.07EE1793 pKa = 4.56VTKK1796 pKa = 9.79YY1797 pKa = 9.42TLDD1800 pKa = 3.59LAVNPEE1806 pKa = 3.77GSGTVTGAGTYY1817 pKa = 9.49AAGYY1821 pKa = 6.28TVSITATPNEE1831 pKa = 4.02GWEE1834 pKa = 4.05FANWTDD1840 pKa = 3.56GTGATVSSEE1849 pKa = 3.95ATFDD1853 pKa = 3.65YY1854 pKa = 10.73PMPARR1859 pKa = 11.84DD1860 pKa = 3.27TALIANFEE1868 pKa = 4.18PAPPVLTRR1876 pKa = 11.84IDD1878 pKa = 3.7VSPVEE1883 pKa = 4.06AALEE1887 pKa = 4.14VGEE1890 pKa = 4.41TRR1892 pKa = 11.84QFTATGYY1899 pKa = 10.0DD1900 pKa = 3.26QYY1902 pKa = 11.71GNTIPTGEE1910 pKa = 4.37IVWSSTNEE1918 pKa = 3.67AVGTIDD1924 pKa = 3.56ASGMFTALAAGSTEE1938 pKa = 4.47LVAEE1942 pKa = 4.35ANGISGKK1949 pKa = 10.84ANVTVTPAAPVLTSIAVTPEE1969 pKa = 3.84SPALEE1974 pKa = 4.25VGTTQQFTATCYY1986 pKa = 10.15DD1987 pKa = 3.23QYY1989 pKa = 10.7GQTMPEE1995 pKa = 4.14VTVSWSSSNTSVGTIEE2011 pKa = 4.28SNGMFTALTAGSTTVTASAAGISGQTVATVNPAAPAVTRR2050 pKa = 11.84IAVSPLSVTLDD2061 pKa = 3.15IGDD2064 pKa = 3.81TQAFVATCYY2073 pKa = 10.42DD2074 pKa = 3.23QYY2076 pKa = 10.97EE2077 pKa = 4.3NEE2079 pKa = 4.23MPGTSVSWASDD2090 pKa = 3.09STTVGTIDD2098 pKa = 3.17ATGLFTATAEE2108 pKa = 4.42GSATVTATAGDD2119 pKa = 3.37ISGTAEE2125 pKa = 4.01VAVNPAPPVLTSISVSPAAPTIDD2148 pKa = 5.02AGDD2151 pKa = 3.89TQQFTATCYY2160 pKa = 10.15DD2161 pKa = 3.23QYY2163 pKa = 10.7GQTMPEE2169 pKa = 4.14VTVSWSSSNISVGTIEE2185 pKa = 4.36SNGMFTALTAGSTTVTASAEE2205 pKa = 4.63GISGQAVATVNLAAPVITRR2224 pKa = 11.84IAVSPPSVTLDD2235 pKa = 2.95IGDD2238 pKa = 3.74MQTFVATCYY2247 pKa = 10.33DD2248 pKa = 3.28QYY2250 pKa = 10.97EE2251 pKa = 4.31NEE2253 pKa = 4.27MPGTDD2258 pKa = 3.87VSWSSDD2264 pKa = 3.28DD2265 pKa = 3.77TTVGTIDD2272 pKa = 3.19ATGLFTATAEE2282 pKa = 4.42GSATVTATAGEE2293 pKa = 4.26VSGTAEE2299 pKa = 4.15VTVNPAPPVLTSISVSPAAPTIDD2322 pKa = 3.49VGEE2325 pKa = 4.38TQQFTAICYY2334 pKa = 8.49DD2335 pKa = 3.36QYY2337 pKa = 11.41GAPISDD2343 pKa = 3.7VSVIWSSEE2351 pKa = 3.71NEE2353 pKa = 4.28TVGTIDD2359 pKa = 3.19TSGLFTAIEE2368 pKa = 4.17EE2369 pKa = 4.53GATTITASANGISGTATVTVTPAPRR2394 pKa = 11.84VLTTVDD2400 pKa = 3.55VSPATADD2407 pKa = 3.11IAVGEE2412 pKa = 4.54TEE2414 pKa = 4.87QFTATCYY2421 pKa = 10.0DD2422 pKa = 3.37QNGEE2426 pKa = 4.19VMPDD2430 pKa = 3.41VSVTWSSSNEE2440 pKa = 3.82AVGTMTAGGIFTAHH2454 pKa = 6.46SEE2456 pKa = 4.19GATTVTASAEE2466 pKa = 4.32GVSGSASVTVRR2477 pKa = 11.84RR2478 pKa = 11.84VNTAPIAVDD2487 pKa = 4.08DD2488 pKa = 4.83AFTTNIRR2495 pKa = 11.84TQLTVPAPGVLEE2507 pKa = 4.61NDD2509 pKa = 3.41TDD2511 pKa = 4.0SDD2513 pKa = 4.47GDD2515 pKa = 3.91VLTAALVSKK2524 pKa = 10.05PSNGVLTLNTDD2535 pKa = 3.15GSFTYY2540 pKa = 10.01IPKK2543 pKa = 10.72GNFIGTDD2550 pKa = 3.01TFTYY2554 pKa = 9.75KK2555 pKa = 11.02ANDD2558 pKa = 3.53GSLNSSVATVTITVLATNHH2577 pKa = 6.07APIAADD2583 pKa = 3.49DD2584 pKa = 4.3TVTMTQDD2591 pKa = 2.8TTYY2594 pKa = 10.82AAPAPGVLEE2603 pKa = 4.45NDD2605 pKa = 3.26QDD2607 pKa = 4.2PDD2609 pKa = 3.78GDD2611 pKa = 4.1TVTAKK2616 pKa = 10.52LVSKK2620 pKa = 8.85VTYY2623 pKa = 10.57GSLKK2627 pKa = 10.14LKK2629 pKa = 10.46KK2630 pKa = 9.86DD2631 pKa = 3.31GSFTYY2636 pKa = 10.0IPKK2639 pKa = 10.16PGFTGEE2645 pKa = 5.22DD2646 pKa = 3.27SFTYY2650 pKa = 8.69QTSDD2654 pKa = 2.99GKK2656 pKa = 10.94LSSDD2660 pKa = 2.8IATVRR2665 pKa = 11.84ITVEE2669 pKa = 3.93PTAVIPPVADD2679 pKa = 5.57FSASPLGGKK2688 pKa = 10.08APVFVQFTDD2697 pKa = 3.42TSTGTINSWTWTFGDD2712 pKa = 4.18GGTSTEE2718 pKa = 3.93QNPQYY2723 pKa = 10.96KK2724 pKa = 7.94YY2725 pKa = 9.13TRR2727 pKa = 11.84PGTYY2731 pKa = 8.8TVSLTVTGPGGSDD2744 pKa = 2.72TKK2746 pKa = 10.81TVTDD2750 pKa = 4.3YY2751 pKa = 11.3IQVTGKK2757 pKa = 10.39KK2758 pKa = 9.75AA2759 pKa = 2.93
MM1 pKa = 7.74DD2 pKa = 5.2RR3 pKa = 11.84SRR5 pKa = 11.84VIAGAIVILLAVAVLAVPVAAEE27 pKa = 4.09EE28 pKa = 4.54YY29 pKa = 10.88VFVTKK34 pKa = 10.1WGEE37 pKa = 3.93YY38 pKa = 8.58GTGEE42 pKa = 4.23GFKK45 pKa = 10.93DD46 pKa = 3.63PFDD49 pKa = 3.8IAIDD53 pKa = 3.97TAGHH57 pKa = 6.34IYY59 pKa = 8.73ITEE62 pKa = 3.84TDD64 pKa = 3.62ANHH67 pKa = 6.18ILQRR71 pKa = 11.84ILKK74 pKa = 9.12LDD76 pKa = 3.18SSMNFITKK84 pKa = 8.8WGSYY88 pKa = 7.84GTGNGQFNGPQGIAVNAAGNVYY110 pKa = 10.43VADD113 pKa = 4.42TYY115 pKa = 9.83NHH117 pKa = 7.6RR118 pKa = 11.84IQKK121 pKa = 9.75FDD123 pKa = 3.1SSGNLLTKK131 pKa = 9.37WGSWGSGDD139 pKa = 4.14GQFSYY144 pKa = 10.58PDD146 pKa = 3.6SVAVDD151 pKa = 3.35AAGNVYY157 pKa = 10.42VSDD160 pKa = 3.96TNNGRR165 pKa = 11.84IQKK168 pKa = 9.52FDD170 pKa = 3.43SDD172 pKa = 4.0GTFLGKK178 pKa = 9.32WGSWGSGDD186 pKa = 3.38GQLRR190 pKa = 11.84FSQDD194 pKa = 3.26LVVDD198 pKa = 4.21AAGNIYY204 pKa = 9.32VAEE207 pKa = 4.22YY208 pKa = 10.46GNHH211 pKa = 7.16RR212 pKa = 11.84IQKK215 pKa = 9.55FDD217 pKa = 3.87SNGNFLWKK225 pKa = 10.09KK226 pKa = 9.5GSSGSGDD233 pKa = 3.57GQFLSPYY240 pKa = 9.73GITVDD245 pKa = 3.28AAGNVYY251 pKa = 10.41VADD254 pKa = 3.82TWNHH258 pKa = 7.0RR259 pKa = 11.84IQKK262 pKa = 9.56FDD264 pKa = 3.09SSGNFLTKK272 pKa = 9.65WGSRR276 pKa = 11.84GSGNGQFSEE285 pKa = 4.43PFGVAVDD292 pKa = 3.82SAGNVYY298 pKa = 8.42VTEE301 pKa = 4.42RR302 pKa = 11.84GNDD305 pKa = 3.32RR306 pKa = 11.84VQMFAPASALTVTGIVPASGLNTGSVSITDD336 pKa = 3.72LSGTGFANGATVNLTRR352 pKa = 11.84TGEE355 pKa = 4.26TNIVATDD362 pKa = 3.41VSVLSSTQITCTLDD376 pKa = 3.07LTGAVAGSWNVVVTNPDD393 pKa = 3.13NQFATLTDD401 pKa = 3.69GFTVNAEE408 pKa = 3.46NRR410 pKa = 11.84APVAEE415 pKa = 5.2DD416 pKa = 3.09DD417 pKa = 4.41TYY419 pKa = 9.7TTDD422 pKa = 3.77EE423 pKa = 4.57DD424 pKa = 4.22TLLTISAPGVLANDD438 pKa = 3.76NDD440 pKa = 4.41PDD442 pKa = 4.47GDD444 pKa = 4.02ALTAVLVDD452 pKa = 3.99DD453 pKa = 4.8VSNGTLTLNADD464 pKa = 3.48GSFTYY469 pKa = 10.58LPNTGFSDD477 pKa = 3.2IDD479 pKa = 3.77TFTYY483 pKa = 8.55TANDD487 pKa = 3.9GTQSSNVATVSINVNPASAVLTSIVVTPVQPILEE521 pKa = 4.34IGGTQQFTATGYY533 pKa = 10.12DD534 pKa = 3.35QYY536 pKa = 11.15GTPLPTGEE544 pKa = 4.62IVWSSTNTTVGTIDD558 pKa = 3.17ATGLFTATAEE568 pKa = 4.3GSTTVTATAGEE579 pKa = 4.26VSGTAEE585 pKa = 4.1VTVNPALTVTGIVPASGPNTGNVSITNLSGTGFVDD620 pKa = 4.44GATVNLTRR628 pKa = 11.84TGEE631 pKa = 4.26TNIVATDD638 pKa = 3.41VSVLSSTQITCTLDD652 pKa = 3.24LTGAAAGSWNVVVTNPDD669 pKa = 2.55GRR671 pKa = 11.84YY672 pKa = 10.25DD673 pKa = 3.45ILSDD677 pKa = 3.65GFAVVAASEE686 pKa = 4.23IVTFNDD692 pKa = 3.24PNLEE696 pKa = 4.03ATVRR700 pKa = 11.84GALGKK705 pKa = 9.62PVGDD709 pKa = 3.48ITADD713 pKa = 3.44DD714 pKa = 3.87MATFITLGADD724 pKa = 2.75WRR726 pKa = 11.84GIRR729 pKa = 11.84DD730 pKa = 3.66LSGLEE735 pKa = 3.91YY736 pKa = 10.66AVNLQHH742 pKa = 7.68LYY744 pKa = 10.25LQQNRR749 pKa = 11.84QISDD753 pKa = 4.05LGPLAGLTDD762 pKa = 4.68LQTLDD767 pKa = 3.55LWNNQISDD775 pKa = 4.32LSPLAGLTNLSVLLLGSNQISDD797 pKa = 3.61IGPLAGLTDD806 pKa = 4.23LQRR809 pKa = 11.84LHH811 pKa = 7.56LYY813 pKa = 10.46DD814 pKa = 3.53NQIRR818 pKa = 11.84DD819 pKa = 3.63IGPLAGLTNLWEE831 pKa = 3.94LRR833 pKa = 11.84LYY835 pKa = 10.85NNQIRR840 pKa = 11.84DD841 pKa = 3.57IGPLVANSGLGSGDD855 pKa = 3.48EE856 pKa = 4.86VYY858 pKa = 10.8LQYY861 pKa = 11.34NYY863 pKa = 11.17LDD865 pKa = 4.18LTPGSADD872 pKa = 3.31MNDD875 pKa = 2.83IQTLQSRR882 pKa = 11.84GVYY885 pKa = 8.77VAYY888 pKa = 9.89EE889 pKa = 4.02PQNPVPTYY897 pKa = 10.22TLDD900 pKa = 3.64LAVNPEE906 pKa = 3.93GGGTVTGAGTYY917 pKa = 9.84KK918 pKa = 10.78AGDD921 pKa = 3.91TVPITATPNEE931 pKa = 3.83GWEE934 pKa = 4.21FVNWTNEE941 pKa = 3.44TGATVSSEE949 pKa = 4.17PNFDD953 pKa = 3.5YY954 pKa = 10.84PMPEE958 pKa = 3.72GDD960 pKa = 3.79VALTANFEE968 pKa = 4.47GEE970 pKa = 4.52AITYY974 pKa = 7.91TLGLAVNPAGSGTVTGAGTYY994 pKa = 10.21AAGDD998 pKa = 4.11TVSVTATPNDD1008 pKa = 3.93GYY1010 pKa = 9.63TFVNWTDD1017 pKa = 3.45EE1018 pKa = 4.19TDD1020 pKa = 3.76VTVSTAASFDD1030 pKa = 3.5YY1031 pKa = 10.74TMPDD1035 pKa = 2.86GDD1037 pKa = 3.87VTLTANFAAITYY1049 pKa = 8.43TLDD1052 pKa = 3.49LAVNPEE1058 pKa = 3.99GSGTATGAGTYY1069 pKa = 10.09AAGATVPITATPNEE1083 pKa = 4.08GWEE1086 pKa = 4.05FANWTDD1092 pKa = 3.56GTGATVSSEE1101 pKa = 3.8ATFDD1105 pKa = 3.54YY1106 pKa = 10.86TMPAQEE1112 pKa = 4.32TALTANFEE1120 pKa = 4.16PAPPVLARR1128 pKa = 11.84IEE1130 pKa = 4.1VSPAEE1135 pKa = 4.01ATLEE1139 pKa = 4.14VGATRR1144 pKa = 11.84QFTATGYY1151 pKa = 10.08DD1152 pKa = 3.37QYY1154 pKa = 11.8GNPLSVGEE1162 pKa = 4.53IVWSSTNEE1170 pKa = 3.67AVGTIDD1176 pKa = 3.56ASGMFTALAAGSTEE1190 pKa = 4.47LVAEE1194 pKa = 4.35ANGISGKK1201 pKa = 10.84ANVTVTPAAPVLTSIAVTPEE1221 pKa = 3.85SPALEE1226 pKa = 4.43IGTTQQFTATCYY1238 pKa = 10.15DD1239 pKa = 3.23QYY1241 pKa = 10.97GQTMPDD1247 pKa = 3.31MAVSWSSSNTSVGTIEE1263 pKa = 4.28SNGMFTALTAGSTTVTASAAGVSGQTVATVNPAAPVVTRR1302 pKa = 11.84IAVSPPSVTLDD1313 pKa = 3.29IGDD1316 pKa = 3.81TQAFVATCYY1325 pKa = 10.42DD1326 pKa = 3.23QYY1328 pKa = 10.97EE1329 pKa = 4.3NEE1331 pKa = 4.23MPGTSVSWASDD1342 pKa = 3.09STTVGTIDD1350 pKa = 3.17ATGLFTATAEE1360 pKa = 4.3GSTTVTATAGDD1371 pKa = 3.63VSGTAEE1377 pKa = 4.03VRR1379 pKa = 11.84VNPALTVTGIVPASGPNTGPVSITDD1404 pKa = 3.71LSGTGFADD1412 pKa = 4.16GATVKK1417 pKa = 9.63LTRR1420 pKa = 11.84DD1421 pKa = 3.34GEE1423 pKa = 4.18ADD1425 pKa = 3.15IVATDD1430 pKa = 3.64VAVLSPTQITCTLDD1444 pKa = 3.16LTGAAVGTWNVVVTNPDD1461 pKa = 2.99EE1462 pKa = 4.45QYY1464 pKa = 11.29DD1465 pKa = 3.73ILSDD1469 pKa = 3.81GFAVVAASEE1478 pKa = 4.52VVTFNDD1484 pKa = 3.69PNLEE1488 pKa = 3.85AAVRR1492 pKa = 11.84QEE1494 pKa = 3.65LSKK1497 pKa = 11.07PKK1499 pKa = 10.75GDD1501 pKa = 3.28ITADD1505 pKa = 3.42DD1506 pKa = 4.1MATLTRR1512 pKa = 11.84LSAAGRR1518 pKa = 11.84GIRR1521 pKa = 11.84DD1522 pKa = 3.53LSGLEE1527 pKa = 4.0YY1528 pKa = 10.64AVNLQTLYY1536 pKa = 10.51ISNNQISDD1544 pKa = 4.09LSPLAGLTNLQTLWLQDD1561 pKa = 3.53NQVSDD1566 pKa = 4.43LSPLAGLTNLQRR1578 pKa = 11.84LWLNQNQIRR1587 pKa = 11.84DD1588 pKa = 3.78VSPLAGLTNLRR1599 pKa = 11.84EE1600 pKa = 4.02LLLAVNQISDD1610 pKa = 4.26LSPLAGLTNLGYY1622 pKa = 10.21VQLYY1626 pKa = 9.85RR1627 pKa = 11.84NQISDD1632 pKa = 4.26LSPLAGLTNLGYY1644 pKa = 10.21VQLYY1648 pKa = 9.85RR1649 pKa = 11.84NQISDD1654 pKa = 4.26LSPLAGLTNLYY1665 pKa = 10.49FLDD1668 pKa = 3.21ISYY1671 pKa = 10.43NQISDD1676 pKa = 4.26LSPLASLTNLYY1687 pKa = 10.55FLDD1690 pKa = 3.21ISYY1693 pKa = 10.4NQISDD1698 pKa = 3.68ISPLAGLTRR1707 pKa = 11.84LSRR1710 pKa = 11.84LSLDD1714 pKa = 3.46NNQISDD1720 pKa = 3.8ISPLAGLINLYY1731 pKa = 10.12VLNLNYY1737 pKa = 10.58NQIRR1741 pKa = 11.84DD1742 pKa = 3.76ISPLVANSGLAGDD1755 pKa = 5.2DD1756 pKa = 3.85VYY1758 pKa = 11.61LQYY1761 pKa = 11.36NYY1763 pKa = 11.1LDD1765 pKa = 4.08LTPGSAAMNDD1775 pKa = 3.04IQTLQSRR1782 pKa = 11.84GVYY1785 pKa = 9.2VVYY1788 pKa = 10.25EE1789 pKa = 4.13PQHH1792 pKa = 6.07EE1793 pKa = 4.56VTKK1796 pKa = 9.79YY1797 pKa = 9.42TLDD1800 pKa = 3.59LAVNPEE1806 pKa = 3.77GSGTVTGAGTYY1817 pKa = 9.49AAGYY1821 pKa = 6.28TVSITATPNEE1831 pKa = 4.02GWEE1834 pKa = 4.05FANWTDD1840 pKa = 3.56GTGATVSSEE1849 pKa = 3.95ATFDD1853 pKa = 3.65YY1854 pKa = 10.73PMPARR1859 pKa = 11.84DD1860 pKa = 3.27TALIANFEE1868 pKa = 4.18PAPPVLTRR1876 pKa = 11.84IDD1878 pKa = 3.7VSPVEE1883 pKa = 4.06AALEE1887 pKa = 4.14VGEE1890 pKa = 4.41TRR1892 pKa = 11.84QFTATGYY1899 pKa = 10.0DD1900 pKa = 3.26QYY1902 pKa = 11.71GNTIPTGEE1910 pKa = 4.37IVWSSTNEE1918 pKa = 3.67AVGTIDD1924 pKa = 3.56ASGMFTALAAGSTEE1938 pKa = 4.47LVAEE1942 pKa = 4.35ANGISGKK1949 pKa = 10.84ANVTVTPAAPVLTSIAVTPEE1969 pKa = 3.84SPALEE1974 pKa = 4.25VGTTQQFTATCYY1986 pKa = 10.15DD1987 pKa = 3.23QYY1989 pKa = 10.7GQTMPEE1995 pKa = 4.14VTVSWSSSNTSVGTIEE2011 pKa = 4.28SNGMFTALTAGSTTVTASAAGISGQTVATVNPAAPAVTRR2050 pKa = 11.84IAVSPLSVTLDD2061 pKa = 3.15IGDD2064 pKa = 3.81TQAFVATCYY2073 pKa = 10.42DD2074 pKa = 3.23QYY2076 pKa = 10.97EE2077 pKa = 4.3NEE2079 pKa = 4.23MPGTSVSWASDD2090 pKa = 3.09STTVGTIDD2098 pKa = 3.17ATGLFTATAEE2108 pKa = 4.42GSATVTATAGDD2119 pKa = 3.37ISGTAEE2125 pKa = 4.01VAVNPAPPVLTSISVSPAAPTIDD2148 pKa = 5.02AGDD2151 pKa = 3.89TQQFTATCYY2160 pKa = 10.15DD2161 pKa = 3.23QYY2163 pKa = 10.7GQTMPEE2169 pKa = 4.14VTVSWSSSNISVGTIEE2185 pKa = 4.36SNGMFTALTAGSTTVTASAEE2205 pKa = 4.63GISGQAVATVNLAAPVITRR2224 pKa = 11.84IAVSPPSVTLDD2235 pKa = 2.95IGDD2238 pKa = 3.74MQTFVATCYY2247 pKa = 10.33DD2248 pKa = 3.28QYY2250 pKa = 10.97EE2251 pKa = 4.31NEE2253 pKa = 4.27MPGTDD2258 pKa = 3.87VSWSSDD2264 pKa = 3.28DD2265 pKa = 3.77TTVGTIDD2272 pKa = 3.19ATGLFTATAEE2282 pKa = 4.42GSATVTATAGEE2293 pKa = 4.26VSGTAEE2299 pKa = 4.15VTVNPAPPVLTSISVSPAAPTIDD2322 pKa = 3.49VGEE2325 pKa = 4.38TQQFTAICYY2334 pKa = 8.49DD2335 pKa = 3.36QYY2337 pKa = 11.41GAPISDD2343 pKa = 3.7VSVIWSSEE2351 pKa = 3.71NEE2353 pKa = 4.28TVGTIDD2359 pKa = 3.19TSGLFTAIEE2368 pKa = 4.17EE2369 pKa = 4.53GATTITASANGISGTATVTVTPAPRR2394 pKa = 11.84VLTTVDD2400 pKa = 3.55VSPATADD2407 pKa = 3.11IAVGEE2412 pKa = 4.54TEE2414 pKa = 4.87QFTATCYY2421 pKa = 10.0DD2422 pKa = 3.37QNGEE2426 pKa = 4.19VMPDD2430 pKa = 3.41VSVTWSSSNEE2440 pKa = 3.82AVGTMTAGGIFTAHH2454 pKa = 6.46SEE2456 pKa = 4.19GATTVTASAEE2466 pKa = 4.32GVSGSASVTVRR2477 pKa = 11.84RR2478 pKa = 11.84VNTAPIAVDD2487 pKa = 4.08DD2488 pKa = 4.83AFTTNIRR2495 pKa = 11.84TQLTVPAPGVLEE2507 pKa = 4.61NDD2509 pKa = 3.41TDD2511 pKa = 4.0SDD2513 pKa = 4.47GDD2515 pKa = 3.91VLTAALVSKK2524 pKa = 10.05PSNGVLTLNTDD2535 pKa = 3.15GSFTYY2540 pKa = 10.01IPKK2543 pKa = 10.72GNFIGTDD2550 pKa = 3.01TFTYY2554 pKa = 9.75KK2555 pKa = 11.02ANDD2558 pKa = 3.53GSLNSSVATVTITVLATNHH2577 pKa = 6.07APIAADD2583 pKa = 3.49DD2584 pKa = 4.3TVTMTQDD2591 pKa = 2.8TTYY2594 pKa = 10.82AAPAPGVLEE2603 pKa = 4.45NDD2605 pKa = 3.26QDD2607 pKa = 4.2PDD2609 pKa = 3.78GDD2611 pKa = 4.1TVTAKK2616 pKa = 10.52LVSKK2620 pKa = 8.85VTYY2623 pKa = 10.57GSLKK2627 pKa = 10.14LKK2629 pKa = 10.46KK2630 pKa = 9.86DD2631 pKa = 3.31GSFTYY2636 pKa = 10.0IPKK2639 pKa = 10.16PGFTGEE2645 pKa = 5.22DD2646 pKa = 3.27SFTYY2650 pKa = 8.69QTSDD2654 pKa = 2.99GKK2656 pKa = 10.94LSSDD2660 pKa = 2.8IATVRR2665 pKa = 11.84ITVEE2669 pKa = 3.93PTAVIPPVADD2679 pKa = 5.57FSASPLGGKK2688 pKa = 10.08APVFVQFTDD2697 pKa = 3.42TSTGTINSWTWTFGDD2712 pKa = 4.18GGTSTEE2718 pKa = 3.93QNPQYY2723 pKa = 10.96KK2724 pKa = 7.94YY2725 pKa = 9.13TRR2727 pKa = 11.84PGTYY2731 pKa = 8.8TVSLTVTGPGGSDD2744 pKa = 2.72TKK2746 pKa = 10.81TVTDD2750 pKa = 4.3YY2751 pKa = 11.3IQVTGKK2757 pKa = 10.39KK2758 pKa = 9.75AA2759 pKa = 2.93
Molecular weight: 285.87 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W6PQI5|W6PQI5_METBM Uncharacterized protein OS=Methanoculleus bourgensis (strain ATCC 43281 / DSM 3045 / OCM 15 / MS2) OX=1201294 GN=BN140_3046 PE=4 SV=1
MM1 pKa = 7.38PTTVHH6 pKa = 6.52CYY8 pKa = 10.18VGEE11 pKa = 4.34TEE13 pKa = 4.73NGPDD17 pKa = 3.95PAPGFAIPDD26 pKa = 4.18FKK28 pKa = 11.3QLMRR32 pKa = 11.84FQQRR36 pKa = 11.84LNLTGIYY43 pKa = 9.77PGIQCIPGHH52 pKa = 5.37VRR54 pKa = 11.84ANGGIPSLPDD64 pKa = 3.13RR65 pKa = 11.84WCPPGSGGRR74 pKa = 11.84TPMHH78 pKa = 6.74AGFGATRR85 pKa = 11.84YY86 pKa = 9.63KK87 pKa = 10.41PRR89 pKa = 11.84RR90 pKa = 11.84QCGGRR95 pKa = 11.84DD96 pKa = 3.49SNRR99 pKa = 11.84KK100 pKa = 8.65FPRR103 pKa = 11.84PRR105 pKa = 11.84APKK108 pKa = 10.46SLDD111 pKa = 3.03IPSRR115 pKa = 3.63
MM1 pKa = 7.38PTTVHH6 pKa = 6.52CYY8 pKa = 10.18VGEE11 pKa = 4.34TEE13 pKa = 4.73NGPDD17 pKa = 3.95PAPGFAIPDD26 pKa = 4.18FKK28 pKa = 11.3QLMRR32 pKa = 11.84FQQRR36 pKa = 11.84LNLTGIYY43 pKa = 9.77PGIQCIPGHH52 pKa = 5.37VRR54 pKa = 11.84ANGGIPSLPDD64 pKa = 3.13RR65 pKa = 11.84WCPPGSGGRR74 pKa = 11.84TPMHH78 pKa = 6.74AGFGATRR85 pKa = 11.84YY86 pKa = 9.63KK87 pKa = 10.41PRR89 pKa = 11.84RR90 pKa = 11.84QCGGRR95 pKa = 11.84DD96 pKa = 3.49SNRR99 pKa = 11.84KK100 pKa = 8.65FPRR103 pKa = 11.84PRR105 pKa = 11.84APKK108 pKa = 10.46SLDD111 pKa = 3.03IPSRR115 pKa = 3.63
Molecular weight: 12.64 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
768769 |
31 |
2759 |
298.6 |
32.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.902 ± 0.065 | 1.314 ± 0.024 |
5.765 ± 0.035 | 6.935 ± 0.055 |
3.542 ± 0.028 | 8.469 ± 0.046 |
1.967 ± 0.024 | 6.335 ± 0.042 |
3.352 ± 0.041 | 9.402 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.408 ± 0.023 | 2.738 ± 0.036 |
5.089 ± 0.035 | 2.508 ± 0.023 |
7.109 ± 0.071 | 5.21 ± 0.04 |
5.679 ± 0.066 | 8.182 ± 0.045 |
1.047 ± 0.02 | 3.048 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
