
Chloroflexus aurantiacus (strain ATCC 29366 / DSM 635 / J-10-fl)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Chloroflexi; Chloroflexia; Chloroflexales; Chloroflexineae; Chloroflexaceae; Chloroflexus; Chloroflexus aurantiacus
Average proteome isoelectric point is 6.43
Get precalculated fractions of proteins
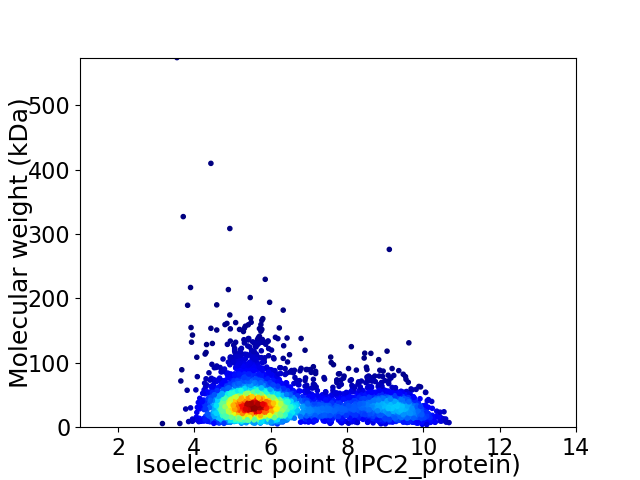
Virtual 2D-PAGE plot for 3850 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A9WFB5|A9WFB5_CHLAA NADH-quinone oxidoreductase E subunit OS=Chloroflexus aurantiacus (strain ATCC 29366 / DSM 635 / J-10-fl) OX=324602 GN=Caur_2900 PE=3 SV=1
MM1 pKa = 7.14TVVPLPRR8 pKa = 11.84PAFPLPLPRR17 pKa = 11.84SPITLLPLSLLLVLLLAAIQPLVVFIAPVLPSTTTPALPYY57 pKa = 10.71ARR59 pKa = 11.84TDD61 pKa = 3.34SNVFIEE67 pKa = 4.15NWGQFDD73 pKa = 3.84PAFVAILIGQDD84 pKa = 2.56ARR86 pKa = 11.84LAVGHH91 pKa = 6.98DD92 pKa = 3.81GRR94 pKa = 11.84LHH96 pKa = 5.68LHH98 pKa = 6.21VDD100 pKa = 4.14GAAPLTFTAAGVTTTPDD117 pKa = 3.17VSLHH121 pKa = 6.22GPLATTISFLTGNDD135 pKa = 3.49PNQHH139 pKa = 5.04VNVPVWEE146 pKa = 4.37VARR149 pKa = 11.84LALTPGVTLEE159 pKa = 4.13LTVIDD164 pKa = 4.53GRR166 pKa = 11.84LRR168 pKa = 11.84LCARR172 pKa = 11.84TADD175 pKa = 3.68PTLLNRR181 pKa = 11.84VQMQVSGGEE190 pKa = 4.13VIGMVDD196 pKa = 4.38GEE198 pKa = 4.9LEE200 pKa = 4.38VQAGAHH206 pKa = 6.26RR207 pKa = 11.84LRR209 pKa = 11.84LPLLEE214 pKa = 4.64AVTPAGIPHH223 pKa = 6.02QQTFSPAIAPDD234 pKa = 3.84GVVRR238 pKa = 11.84APVGIPEE245 pKa = 5.22GIGITGQATPTQTSAAMLLGASTFVGGAAGSGEE278 pKa = 4.22DD279 pKa = 3.46TATAMVIDD287 pKa = 4.37GQGAVYY293 pKa = 10.44VAGNTGSADD302 pKa = 3.42FPTTLGAYY310 pKa = 9.19DD311 pKa = 3.66RR312 pKa = 11.84SLNGGDD318 pKa = 3.57RR319 pKa = 11.84GDD321 pKa = 3.91AFVSRR326 pKa = 11.84LSGDD330 pKa = 3.48LRR332 pKa = 11.84NLLAGTFLGGSDD344 pKa = 3.02WDD346 pKa = 4.01EE347 pKa = 4.77ANDD350 pKa = 4.7LALDD354 pKa = 3.86AQGNVYY360 pKa = 10.26VAGEE364 pKa = 4.13TSSTDD369 pKa = 3.49FPTTLGAYY377 pKa = 9.58DD378 pKa = 3.37RR379 pKa = 11.84TYY381 pKa = 11.33NGDD384 pKa = 3.36GSDD387 pKa = 3.56VFVSRR392 pKa = 11.84LSGDD396 pKa = 3.46LGSLLASTFLGGSYY410 pKa = 9.79WNFAGALALDD420 pKa = 4.33GQGTVYY426 pKa = 10.72VAGGTSSTDD435 pKa = 3.44FPTTLGAYY443 pKa = 9.61DD444 pKa = 3.32RR445 pKa = 11.84TYY447 pKa = 11.38NGGSSDD453 pKa = 3.72VFVSRR458 pKa = 11.84LSGDD462 pKa = 3.85LNSLLASTFLGGSAWDD478 pKa = 3.47SATALVLDD486 pKa = 4.34GQGNVYY492 pKa = 10.25VAGEE496 pKa = 4.19TGSADD501 pKa = 3.79FPTMAGAYY509 pKa = 9.45DD510 pKa = 3.88GSYY513 pKa = 11.25NSGDD517 pKa = 3.28RR518 pKa = 11.84DD519 pKa = 3.54AFASKK524 pKa = 10.77LSGDD528 pKa = 4.02LGSLLTSTFLGGSDD542 pKa = 3.09WDD544 pKa = 3.88VATALALDD552 pKa = 4.21GQGTVYY558 pKa = 10.59VAGYY562 pKa = 8.22TSSTNFPTTLGAYY575 pKa = 9.61DD576 pKa = 3.32RR577 pKa = 11.84TYY579 pKa = 11.57NGGSSDD585 pKa = 3.42AFAGRR590 pKa = 11.84LSGDD594 pKa = 3.48LGSLLTSTFLGGNDD608 pKa = 3.2SEE610 pKa = 4.83YY611 pKa = 11.38ALSLALDD618 pKa = 4.02GQGAVYY624 pKa = 11.08VMGEE628 pKa = 4.07TGSTIFPTTAGAYY641 pKa = 9.85DD642 pKa = 3.3RR643 pKa = 11.84VYY645 pKa = 11.18NGGYY649 pKa = 9.39RR650 pKa = 11.84DD651 pKa = 3.74AFVSRR656 pKa = 11.84LSSNLGSLLASTFLGGSGWDD676 pKa = 3.29VAIALTLDD684 pKa = 3.72GQGTVYY690 pKa = 10.59VAGYY694 pKa = 8.22TSSTNFPTTLGAYY707 pKa = 9.64DD708 pKa = 3.91GSYY711 pKa = 10.99NGGDD715 pKa = 3.13RR716 pKa = 11.84DD717 pKa = 3.81AFASKK722 pKa = 10.77LSGDD726 pKa = 4.02LGSLLTSTFLGGMFGNGNDD745 pKa = 3.65VANSLAISSQDD756 pKa = 3.28TVYY759 pKa = 10.77VAGWTGSANFPTTAGAYY776 pKa = 10.06DD777 pKa = 3.68GVFNRR782 pKa = 11.84GFRR785 pKa = 11.84DD786 pKa = 3.41AFVSRR791 pKa = 11.84LSSDD795 pKa = 3.31LGSLLASTFLGGSDD809 pKa = 3.09WDD811 pKa = 3.78VATALVLDD819 pKa = 4.39GQGNVYY825 pKa = 10.24VAGEE829 pKa = 4.06TWSANFPTTAGAYY842 pKa = 9.48DD843 pKa = 3.34RR844 pKa = 11.84TYY846 pKa = 11.51NGGARR851 pKa = 11.84DD852 pKa = 3.71AFVSKK857 pKa = 10.88LSGNLGSLLASTFLGGSDD875 pKa = 3.36WDD877 pKa = 3.91SATALALDD885 pKa = 4.06GQGNVYY891 pKa = 10.31VAGDD895 pKa = 3.77TYY897 pKa = 11.17STDD900 pKa = 3.7FPTTAGVYY908 pKa = 10.03DD909 pKa = 3.69RR910 pKa = 11.84AYY912 pKa = 11.08NGGGSDD918 pKa = 3.54TFVSRR923 pKa = 11.84LSGDD927 pKa = 3.2LGGLLAGSFLGGSLPDD943 pKa = 3.45SATALALDD951 pKa = 4.06GQGNVYY957 pKa = 9.68VAGWTDD963 pKa = 3.32SANFPTTAGSYY974 pKa = 10.34DD975 pKa = 3.28GSFNGSDD982 pKa = 3.46GFVSKK987 pKa = 10.95LSGEE991 pKa = 4.1LRR993 pKa = 11.84SLVASTFLGGSIDD1006 pKa = 3.85EE1007 pKa = 4.6SATALTLDD1015 pKa = 3.75GQGNVYY1021 pKa = 10.25VAGEE1025 pKa = 4.13TSSTDD1030 pKa = 3.49FPTTLGAHH1038 pKa = 6.45DD1039 pKa = 3.52RR1040 pKa = 11.84TYY1042 pKa = 11.44NGGGSDD1048 pKa = 3.64VFVSRR1053 pKa = 11.84LSGDD1057 pKa = 3.55LGSQLASTFLGGSDD1071 pKa = 3.36WDD1073 pKa = 3.82SATALVLDD1081 pKa = 4.58GQGSVYY1087 pKa = 10.62VAGEE1091 pKa = 4.05TGSADD1096 pKa = 3.79FPTMAGAYY1104 pKa = 9.12DD1105 pKa = 3.95GSYY1108 pKa = 10.44NGSGDD1113 pKa = 3.7TFVSRR1118 pKa = 11.84LSGNLGGLLASTFLGGSAWDD1138 pKa = 3.57SATALALDD1146 pKa = 4.19GQGTVYY1152 pKa = 10.56VAGYY1156 pKa = 8.05TEE1158 pKa = 5.01SNNFPITAGAYY1169 pKa = 8.54GGVYY1173 pKa = 10.53NGDD1176 pKa = 2.95RR1177 pKa = 11.84DD1178 pKa = 3.98AFVSKK1183 pKa = 10.67LVFAFTKK1190 pKa = 9.91TSSAVSTTNQSINLTLQWQSAGATVHH1216 pKa = 6.78HH1217 pKa = 6.36YY1218 pKa = 10.13RR1219 pKa = 11.84YY1220 pKa = 10.48CLDD1223 pKa = 3.84TNPGCTPTTNVGANTSVTVAGITPNTTYY1251 pKa = 10.07YY1252 pKa = 8.51WQVRR1256 pKa = 11.84ACADD1260 pKa = 3.21SDD1262 pKa = 4.09CAVFIDD1268 pKa = 5.13ANNGQHH1274 pKa = 6.36WSLKK1278 pKa = 7.49TSFMIYY1284 pKa = 9.1LTFAARR1290 pKa = 3.46
MM1 pKa = 7.14TVVPLPRR8 pKa = 11.84PAFPLPLPRR17 pKa = 11.84SPITLLPLSLLLVLLLAAIQPLVVFIAPVLPSTTTPALPYY57 pKa = 10.71ARR59 pKa = 11.84TDD61 pKa = 3.34SNVFIEE67 pKa = 4.15NWGQFDD73 pKa = 3.84PAFVAILIGQDD84 pKa = 2.56ARR86 pKa = 11.84LAVGHH91 pKa = 6.98DD92 pKa = 3.81GRR94 pKa = 11.84LHH96 pKa = 5.68LHH98 pKa = 6.21VDD100 pKa = 4.14GAAPLTFTAAGVTTTPDD117 pKa = 3.17VSLHH121 pKa = 6.22GPLATTISFLTGNDD135 pKa = 3.49PNQHH139 pKa = 5.04VNVPVWEE146 pKa = 4.37VARR149 pKa = 11.84LALTPGVTLEE159 pKa = 4.13LTVIDD164 pKa = 4.53GRR166 pKa = 11.84LRR168 pKa = 11.84LCARR172 pKa = 11.84TADD175 pKa = 3.68PTLLNRR181 pKa = 11.84VQMQVSGGEE190 pKa = 4.13VIGMVDD196 pKa = 4.38GEE198 pKa = 4.9LEE200 pKa = 4.38VQAGAHH206 pKa = 6.26RR207 pKa = 11.84LRR209 pKa = 11.84LPLLEE214 pKa = 4.64AVTPAGIPHH223 pKa = 6.02QQTFSPAIAPDD234 pKa = 3.84GVVRR238 pKa = 11.84APVGIPEE245 pKa = 5.22GIGITGQATPTQTSAAMLLGASTFVGGAAGSGEE278 pKa = 4.22DD279 pKa = 3.46TATAMVIDD287 pKa = 4.37GQGAVYY293 pKa = 10.44VAGNTGSADD302 pKa = 3.42FPTTLGAYY310 pKa = 9.19DD311 pKa = 3.66RR312 pKa = 11.84SLNGGDD318 pKa = 3.57RR319 pKa = 11.84GDD321 pKa = 3.91AFVSRR326 pKa = 11.84LSGDD330 pKa = 3.48LRR332 pKa = 11.84NLLAGTFLGGSDD344 pKa = 3.02WDD346 pKa = 4.01EE347 pKa = 4.77ANDD350 pKa = 4.7LALDD354 pKa = 3.86AQGNVYY360 pKa = 10.26VAGEE364 pKa = 4.13TSSTDD369 pKa = 3.49FPTTLGAYY377 pKa = 9.58DD378 pKa = 3.37RR379 pKa = 11.84TYY381 pKa = 11.33NGDD384 pKa = 3.36GSDD387 pKa = 3.56VFVSRR392 pKa = 11.84LSGDD396 pKa = 3.46LGSLLASTFLGGSYY410 pKa = 9.79WNFAGALALDD420 pKa = 4.33GQGTVYY426 pKa = 10.72VAGGTSSTDD435 pKa = 3.44FPTTLGAYY443 pKa = 9.61DD444 pKa = 3.32RR445 pKa = 11.84TYY447 pKa = 11.38NGGSSDD453 pKa = 3.72VFVSRR458 pKa = 11.84LSGDD462 pKa = 3.85LNSLLASTFLGGSAWDD478 pKa = 3.47SATALVLDD486 pKa = 4.34GQGNVYY492 pKa = 10.25VAGEE496 pKa = 4.19TGSADD501 pKa = 3.79FPTMAGAYY509 pKa = 9.45DD510 pKa = 3.88GSYY513 pKa = 11.25NSGDD517 pKa = 3.28RR518 pKa = 11.84DD519 pKa = 3.54AFASKK524 pKa = 10.77LSGDD528 pKa = 4.02LGSLLTSTFLGGSDD542 pKa = 3.09WDD544 pKa = 3.88VATALALDD552 pKa = 4.21GQGTVYY558 pKa = 10.59VAGYY562 pKa = 8.22TSSTNFPTTLGAYY575 pKa = 9.61DD576 pKa = 3.32RR577 pKa = 11.84TYY579 pKa = 11.57NGGSSDD585 pKa = 3.42AFAGRR590 pKa = 11.84LSGDD594 pKa = 3.48LGSLLTSTFLGGNDD608 pKa = 3.2SEE610 pKa = 4.83YY611 pKa = 11.38ALSLALDD618 pKa = 4.02GQGAVYY624 pKa = 11.08VMGEE628 pKa = 4.07TGSTIFPTTAGAYY641 pKa = 9.85DD642 pKa = 3.3RR643 pKa = 11.84VYY645 pKa = 11.18NGGYY649 pKa = 9.39RR650 pKa = 11.84DD651 pKa = 3.74AFVSRR656 pKa = 11.84LSSNLGSLLASTFLGGSGWDD676 pKa = 3.29VAIALTLDD684 pKa = 3.72GQGTVYY690 pKa = 10.59VAGYY694 pKa = 8.22TSSTNFPTTLGAYY707 pKa = 9.64DD708 pKa = 3.91GSYY711 pKa = 10.99NGGDD715 pKa = 3.13RR716 pKa = 11.84DD717 pKa = 3.81AFASKK722 pKa = 10.77LSGDD726 pKa = 4.02LGSLLTSTFLGGMFGNGNDD745 pKa = 3.65VANSLAISSQDD756 pKa = 3.28TVYY759 pKa = 10.77VAGWTGSANFPTTAGAYY776 pKa = 10.06DD777 pKa = 3.68GVFNRR782 pKa = 11.84GFRR785 pKa = 11.84DD786 pKa = 3.41AFVSRR791 pKa = 11.84LSSDD795 pKa = 3.31LGSLLASTFLGGSDD809 pKa = 3.09WDD811 pKa = 3.78VATALVLDD819 pKa = 4.39GQGNVYY825 pKa = 10.24VAGEE829 pKa = 4.06TWSANFPTTAGAYY842 pKa = 9.48DD843 pKa = 3.34RR844 pKa = 11.84TYY846 pKa = 11.51NGGARR851 pKa = 11.84DD852 pKa = 3.71AFVSKK857 pKa = 10.88LSGNLGSLLASTFLGGSDD875 pKa = 3.36WDD877 pKa = 3.91SATALALDD885 pKa = 4.06GQGNVYY891 pKa = 10.31VAGDD895 pKa = 3.77TYY897 pKa = 11.17STDD900 pKa = 3.7FPTTAGVYY908 pKa = 10.03DD909 pKa = 3.69RR910 pKa = 11.84AYY912 pKa = 11.08NGGGSDD918 pKa = 3.54TFVSRR923 pKa = 11.84LSGDD927 pKa = 3.2LGGLLAGSFLGGSLPDD943 pKa = 3.45SATALALDD951 pKa = 4.06GQGNVYY957 pKa = 9.68VAGWTDD963 pKa = 3.32SANFPTTAGSYY974 pKa = 10.34DD975 pKa = 3.28GSFNGSDD982 pKa = 3.46GFVSKK987 pKa = 10.95LSGEE991 pKa = 4.1LRR993 pKa = 11.84SLVASTFLGGSIDD1006 pKa = 3.85EE1007 pKa = 4.6SATALTLDD1015 pKa = 3.75GQGNVYY1021 pKa = 10.25VAGEE1025 pKa = 4.13TSSTDD1030 pKa = 3.49FPTTLGAHH1038 pKa = 6.45DD1039 pKa = 3.52RR1040 pKa = 11.84TYY1042 pKa = 11.44NGGGSDD1048 pKa = 3.64VFVSRR1053 pKa = 11.84LSGDD1057 pKa = 3.55LGSQLASTFLGGSDD1071 pKa = 3.36WDD1073 pKa = 3.82SATALVLDD1081 pKa = 4.58GQGSVYY1087 pKa = 10.62VAGEE1091 pKa = 4.05TGSADD1096 pKa = 3.79FPTMAGAYY1104 pKa = 9.12DD1105 pKa = 3.95GSYY1108 pKa = 10.44NGSGDD1113 pKa = 3.7TFVSRR1118 pKa = 11.84LSGNLGGLLASTFLGGSAWDD1138 pKa = 3.57SATALALDD1146 pKa = 4.19GQGTVYY1152 pKa = 10.56VAGYY1156 pKa = 8.05TEE1158 pKa = 5.01SNNFPITAGAYY1169 pKa = 8.54GGVYY1173 pKa = 10.53NGDD1176 pKa = 2.95RR1177 pKa = 11.84DD1178 pKa = 3.98AFVSKK1183 pKa = 10.67LVFAFTKK1190 pKa = 9.91TSSAVSTTNQSINLTLQWQSAGATVHH1216 pKa = 6.78HH1217 pKa = 6.36YY1218 pKa = 10.13RR1219 pKa = 11.84YY1220 pKa = 10.48CLDD1223 pKa = 3.84TNPGCTPTTNVGANTSVTVAGITPNTTYY1251 pKa = 10.07YY1252 pKa = 8.51WQVRR1256 pKa = 11.84ACADD1260 pKa = 3.21SDD1262 pKa = 4.09CAVFIDD1268 pKa = 5.13ANNGQHH1274 pKa = 6.36WSLKK1278 pKa = 7.49TSFMIYY1284 pKa = 9.1LTFAARR1290 pKa = 3.46
Molecular weight: 132.07 kDa
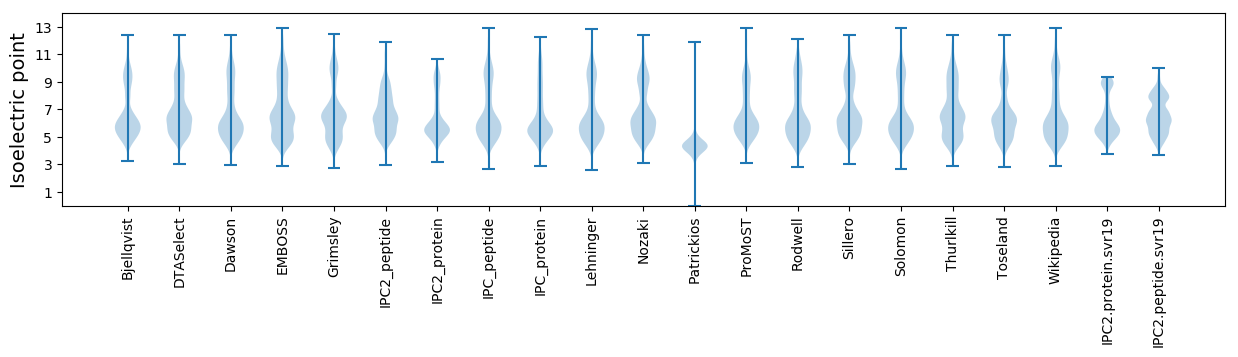
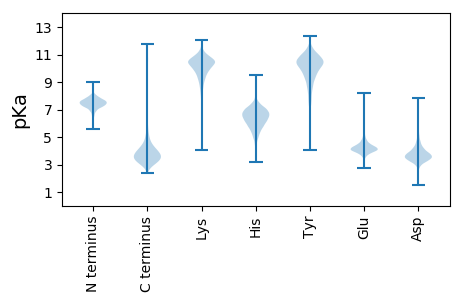
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A9WGV5|A9WGV5_CHLAA Glycosyl hydrolase 53 domain protein OS=Chloroflexus aurantiacus (strain ATCC 29366 / DSM 635 / J-10-fl) OX=324602 GN=Caur_3072 PE=4 SV=1
MM1 pKa = 7.56AANPKK6 pKa = 8.75TATPRR11 pKa = 11.84PRR13 pKa = 11.84HH14 pKa = 4.85VPKK17 pKa = 10.64RR18 pKa = 11.84MCIVCRR24 pKa = 11.84QSDD27 pKa = 3.65AKK29 pKa = 10.43RR30 pKa = 11.84ALIRR34 pKa = 11.84LVRR37 pKa = 11.84NADD40 pKa = 3.08GRR42 pKa = 11.84VVIDD46 pKa = 3.85PTGKK50 pKa = 9.46QPGRR54 pKa = 11.84GAYY57 pKa = 9.77LCRR60 pKa = 11.84NPLCWAQALRR70 pKa = 11.84RR71 pKa = 11.84NALEE75 pKa = 4.06RR76 pKa = 11.84ALKK79 pKa = 10.21IATLHH84 pKa = 6.39PDD86 pKa = 3.05DD87 pKa = 4.58RR88 pKa = 11.84AVIEE92 pKa = 5.09AMGQQLLQEE101 pKa = 4.49TEE103 pKa = 4.29STASAA108 pKa = 3.69
MM1 pKa = 7.56AANPKK6 pKa = 8.75TATPRR11 pKa = 11.84PRR13 pKa = 11.84HH14 pKa = 4.85VPKK17 pKa = 10.64RR18 pKa = 11.84MCIVCRR24 pKa = 11.84QSDD27 pKa = 3.65AKK29 pKa = 10.43RR30 pKa = 11.84ALIRR34 pKa = 11.84LVRR37 pKa = 11.84NADD40 pKa = 3.08GRR42 pKa = 11.84VVIDD46 pKa = 3.85PTGKK50 pKa = 9.46QPGRR54 pKa = 11.84GAYY57 pKa = 9.77LCRR60 pKa = 11.84NPLCWAQALRR70 pKa = 11.84RR71 pKa = 11.84NALEE75 pKa = 4.06RR76 pKa = 11.84ALKK79 pKa = 10.21IATLHH84 pKa = 6.39PDD86 pKa = 3.05DD87 pKa = 4.58RR88 pKa = 11.84AVIEE92 pKa = 5.09AMGQQLLQEE101 pKa = 4.49TEE103 pKa = 4.29STASAA108 pKa = 3.69
Molecular weight: 11.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1405705 |
36 |
5505 |
365.1 |
40.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.275 ± 0.054 | 0.818 ± 0.011 |
5.119 ± 0.035 | 5.401 ± 0.039 |
3.4 ± 0.025 | 7.633 ± 0.042 |
2.124 ± 0.021 | 5.943 ± 0.032 |
1.781 ± 0.026 | 11.305 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.006 ± 0.015 | 2.732 ± 0.032 |
5.909 ± 0.033 | 4.226 ± 0.028 |
7.585 ± 0.041 | 4.928 ± 0.033 |
6.009 ± 0.052 | 7.499 ± 0.03 |
1.595 ± 0.019 | 2.712 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
