
Human papillomavirus 134
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus; Gammapapillomavirus 7
Average proteome isoelectric point is 6.47
Get precalculated fractions of proteins
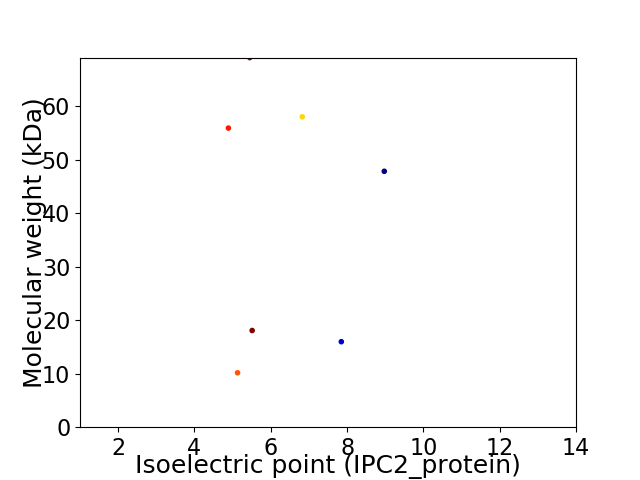
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
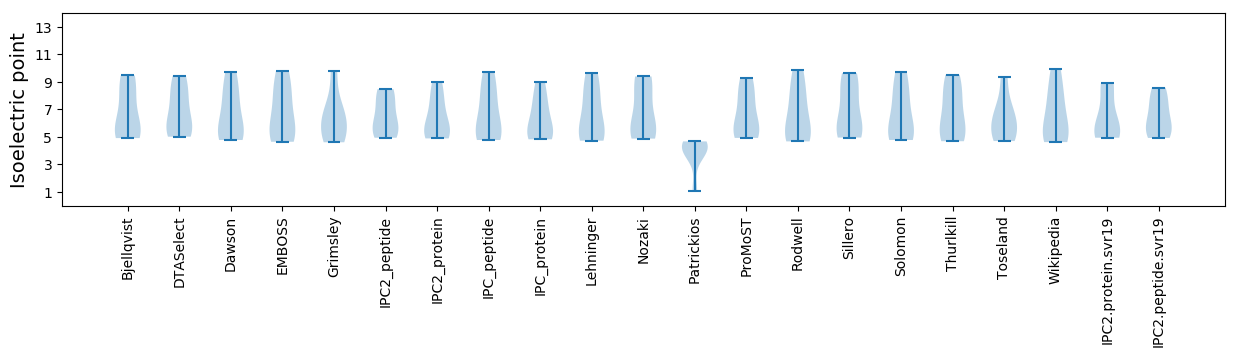
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E7BQB3|E7BQB3_9PAPI Major capsid protein L1 OS=Human papillomavirus 134 OX=909333 GN=L1 PE=3 SV=1
MM1 pKa = 7.55SEE3 pKa = 4.18RR4 pKa = 11.84PRR6 pKa = 11.84KK7 pKa = 7.84RR8 pKa = 11.84TKK10 pKa = 10.15RR11 pKa = 11.84DD12 pKa = 3.18SVTNLYY18 pKa = 8.66NQCKK22 pKa = 9.99ISGSCPDD29 pKa = 3.53DD30 pKa = 3.52VKK32 pKa = 11.61NKK34 pKa = 9.99VEE36 pKa = 4.16EE37 pKa = 4.12TTLADD42 pKa = 3.73RR43 pKa = 11.84LLKK46 pKa = 10.35IFSSLVYY53 pKa = 10.23FGGLGIGTGKK63 pKa = 10.9GIGGSTGYY71 pKa = 10.26RR72 pKa = 11.84PLGGGSGRR80 pKa = 11.84VTAGGTVIRR89 pKa = 11.84PNVAVDD95 pKa = 3.43PLGPTDD101 pKa = 4.21IVPIDD106 pKa = 4.33SISPGTSSIVPLIEE120 pKa = 3.79VTPEE124 pKa = 3.72VVPEE128 pKa = 4.27LEE130 pKa = 4.19AGVINTGFDD139 pKa = 2.91ITTEE143 pKa = 3.8PDD145 pKa = 3.09IIEE148 pKa = 4.34ISSGSNTPAVSTIDD162 pKa = 3.25DD163 pKa = 3.61TAAVLEE169 pKa = 4.49VQPSTTTPRR178 pKa = 11.84RR179 pKa = 11.84VTTSKK184 pKa = 9.42FTNPTYY190 pKa = 8.44MTVVTEE196 pKa = 4.12STITPEE202 pKa = 4.07VSSTAGVFIDD212 pKa = 4.26GALGGDD218 pKa = 3.4VVGEE222 pKa = 4.27NIPLDD227 pKa = 3.89TFNEE231 pKa = 4.01PLEE234 pKa = 4.05FEE236 pKa = 4.3ILEE239 pKa = 4.13EE240 pKa = 4.03QQPKK244 pKa = 9.58TSTPIEE250 pKa = 4.2NVTRR254 pKa = 11.84ALGRR258 pKa = 11.84ARR260 pKa = 11.84EE261 pKa = 4.15LYY263 pKa = 9.51NRR265 pKa = 11.84RR266 pKa = 11.84VRR268 pKa = 11.84QVMTRR273 pKa = 11.84NPNFLTRR280 pKa = 11.84APQAVQFVFEE290 pKa = 4.94NPAFTNDD297 pKa = 2.87VTLTFEE303 pKa = 4.21QDD305 pKa = 3.08LNQIATAAPDD315 pKa = 3.96PDD317 pKa = 3.58FADD320 pKa = 4.07IIKK323 pKa = 10.57LEE325 pKa = 4.03RR326 pKa = 11.84PRR328 pKa = 11.84FSEE331 pKa = 4.06TAEE334 pKa = 3.98GTVRR338 pKa = 11.84LSRR341 pKa = 11.84LGTRR345 pKa = 11.84GTIRR349 pKa = 11.84TRR351 pKa = 11.84SGTQIGEE358 pKa = 4.16RR359 pKa = 11.84VHH361 pKa = 7.15FYY363 pKa = 11.36YY364 pKa = 10.73DD365 pKa = 3.25LSKK368 pKa = 10.72IEE370 pKa = 3.94NSEE373 pKa = 4.22AIEE376 pKa = 3.97LSVLGEE382 pKa = 4.04HH383 pKa = 6.78SGDD386 pKa = 3.12ATFINPLAEE395 pKa = 4.27STFVDD400 pKa = 4.2AEE402 pKa = 4.28NTNMPAIFPEE412 pKa = 4.23EE413 pKa = 4.03EE414 pKa = 4.14LVDD417 pKa = 5.0EE418 pKa = 4.24ITEE421 pKa = 4.33DD422 pKa = 3.82FSNSHH427 pKa = 5.45VVLINGSRR435 pKa = 11.84RR436 pKa = 11.84STMNVPTLPPGVALRR451 pKa = 11.84VYY453 pKa = 10.67VDD455 pKa = 4.62DD456 pKa = 4.2YY457 pKa = 12.01GEE459 pKa = 4.31GLFVSHH465 pKa = 6.39PQGRR469 pKa = 11.84EE470 pKa = 3.44LPAIITPGNAGRR482 pKa = 11.84PSILIDD488 pKa = 3.97DD489 pKa = 4.84FASNDD494 pKa = 3.68FVLHH498 pKa = 6.35PSLNRR503 pKa = 11.84KK504 pKa = 7.77KK505 pKa = 10.27RR506 pKa = 11.84KK507 pKa = 8.98RR508 pKa = 11.84KK509 pKa = 8.79QVHH512 pKa = 5.56FLL514 pKa = 3.45
MM1 pKa = 7.55SEE3 pKa = 4.18RR4 pKa = 11.84PRR6 pKa = 11.84KK7 pKa = 7.84RR8 pKa = 11.84TKK10 pKa = 10.15RR11 pKa = 11.84DD12 pKa = 3.18SVTNLYY18 pKa = 8.66NQCKK22 pKa = 9.99ISGSCPDD29 pKa = 3.53DD30 pKa = 3.52VKK32 pKa = 11.61NKK34 pKa = 9.99VEE36 pKa = 4.16EE37 pKa = 4.12TTLADD42 pKa = 3.73RR43 pKa = 11.84LLKK46 pKa = 10.35IFSSLVYY53 pKa = 10.23FGGLGIGTGKK63 pKa = 10.9GIGGSTGYY71 pKa = 10.26RR72 pKa = 11.84PLGGGSGRR80 pKa = 11.84VTAGGTVIRR89 pKa = 11.84PNVAVDD95 pKa = 3.43PLGPTDD101 pKa = 4.21IVPIDD106 pKa = 4.33SISPGTSSIVPLIEE120 pKa = 3.79VTPEE124 pKa = 3.72VVPEE128 pKa = 4.27LEE130 pKa = 4.19AGVINTGFDD139 pKa = 2.91ITTEE143 pKa = 3.8PDD145 pKa = 3.09IIEE148 pKa = 4.34ISSGSNTPAVSTIDD162 pKa = 3.25DD163 pKa = 3.61TAAVLEE169 pKa = 4.49VQPSTTTPRR178 pKa = 11.84RR179 pKa = 11.84VTTSKK184 pKa = 9.42FTNPTYY190 pKa = 8.44MTVVTEE196 pKa = 4.12STITPEE202 pKa = 4.07VSSTAGVFIDD212 pKa = 4.26GALGGDD218 pKa = 3.4VVGEE222 pKa = 4.27NIPLDD227 pKa = 3.89TFNEE231 pKa = 4.01PLEE234 pKa = 4.05FEE236 pKa = 4.3ILEE239 pKa = 4.13EE240 pKa = 4.03QQPKK244 pKa = 9.58TSTPIEE250 pKa = 4.2NVTRR254 pKa = 11.84ALGRR258 pKa = 11.84ARR260 pKa = 11.84EE261 pKa = 4.15LYY263 pKa = 9.51NRR265 pKa = 11.84RR266 pKa = 11.84VRR268 pKa = 11.84QVMTRR273 pKa = 11.84NPNFLTRR280 pKa = 11.84APQAVQFVFEE290 pKa = 4.94NPAFTNDD297 pKa = 2.87VTLTFEE303 pKa = 4.21QDD305 pKa = 3.08LNQIATAAPDD315 pKa = 3.96PDD317 pKa = 3.58FADD320 pKa = 4.07IIKK323 pKa = 10.57LEE325 pKa = 4.03RR326 pKa = 11.84PRR328 pKa = 11.84FSEE331 pKa = 4.06TAEE334 pKa = 3.98GTVRR338 pKa = 11.84LSRR341 pKa = 11.84LGTRR345 pKa = 11.84GTIRR349 pKa = 11.84TRR351 pKa = 11.84SGTQIGEE358 pKa = 4.16RR359 pKa = 11.84VHH361 pKa = 7.15FYY363 pKa = 11.36YY364 pKa = 10.73DD365 pKa = 3.25LSKK368 pKa = 10.72IEE370 pKa = 3.94NSEE373 pKa = 4.22AIEE376 pKa = 3.97LSVLGEE382 pKa = 4.04HH383 pKa = 6.78SGDD386 pKa = 3.12ATFINPLAEE395 pKa = 4.27STFVDD400 pKa = 4.2AEE402 pKa = 4.28NTNMPAIFPEE412 pKa = 4.23EE413 pKa = 4.03EE414 pKa = 4.14LVDD417 pKa = 5.0EE418 pKa = 4.24ITEE421 pKa = 4.33DD422 pKa = 3.82FSNSHH427 pKa = 5.45VVLINGSRR435 pKa = 11.84RR436 pKa = 11.84STMNVPTLPPGVALRR451 pKa = 11.84VYY453 pKa = 10.67VDD455 pKa = 4.62DD456 pKa = 4.2YY457 pKa = 12.01GEE459 pKa = 4.31GLFVSHH465 pKa = 6.39PQGRR469 pKa = 11.84EE470 pKa = 3.44LPAIITPGNAGRR482 pKa = 11.84PSILIDD488 pKa = 3.97DD489 pKa = 4.84FASNDD494 pKa = 3.68FVLHH498 pKa = 6.35PSLNRR503 pKa = 11.84KK504 pKa = 7.77KK505 pKa = 10.27RR506 pKa = 11.84KK507 pKa = 8.98RR508 pKa = 11.84KK509 pKa = 8.79QVHH512 pKa = 5.56FLL514 pKa = 3.45
Molecular weight: 55.88 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E7BQA8|E7BQA8_9PAPI Protein E7 OS=Human papillomavirus 134 OX=909333 GN=E7 PE=3 SV=1
MM1 pKa = 7.4EE2 pKa = 5.23PVYY5 pKa = 10.45SIPALCRR12 pKa = 11.84LCRR15 pKa = 11.84VTIDD19 pKa = 4.11RR20 pKa = 11.84LHH22 pKa = 6.46LPCLFCHH29 pKa = 6.08NVLSLGDD36 pKa = 4.14LYY38 pKa = 11.87SFMFKK43 pKa = 10.63DD44 pKa = 3.64LNVVIRR50 pKa = 11.84GYY52 pKa = 11.04RR53 pKa = 11.84LFACCVNCLLISANYY68 pKa = 9.79EE69 pKa = 3.66MQQYY73 pKa = 7.28YY74 pKa = 9.66QCTANASTVQCLTRR88 pKa = 11.84KK89 pKa = 9.7SLLFLYY95 pKa = 9.14VRR97 pKa = 11.84CDD99 pKa = 3.11VCMKK103 pKa = 10.07PLSATEE109 pKa = 4.81KK110 pKa = 10.47FDD112 pKa = 4.85CLTKK116 pKa = 10.31NDD118 pKa = 3.73EE119 pKa = 4.09MHH121 pKa = 6.94LVRR124 pKa = 11.84SIWRR128 pKa = 11.84GTCRR132 pKa = 11.84LCRR135 pKa = 11.84KK136 pKa = 9.34KK137 pKa = 10.8
MM1 pKa = 7.4EE2 pKa = 5.23PVYY5 pKa = 10.45SIPALCRR12 pKa = 11.84LCRR15 pKa = 11.84VTIDD19 pKa = 4.11RR20 pKa = 11.84LHH22 pKa = 6.46LPCLFCHH29 pKa = 6.08NVLSLGDD36 pKa = 4.14LYY38 pKa = 11.87SFMFKK43 pKa = 10.63DD44 pKa = 3.64LNVVIRR50 pKa = 11.84GYY52 pKa = 11.04RR53 pKa = 11.84LFACCVNCLLISANYY68 pKa = 9.79EE69 pKa = 3.66MQQYY73 pKa = 7.28YY74 pKa = 9.66QCTANASTVQCLTRR88 pKa = 11.84KK89 pKa = 9.7SLLFLYY95 pKa = 9.14VRR97 pKa = 11.84CDD99 pKa = 3.11VCMKK103 pKa = 10.07PLSATEE109 pKa = 4.81KK110 pKa = 10.47FDD112 pKa = 4.85CLTKK116 pKa = 10.31NDD118 pKa = 3.73EE119 pKa = 4.09MHH121 pKa = 6.94LVRR124 pKa = 11.84SIWRR128 pKa = 11.84GTCRR132 pKa = 11.84LCRR135 pKa = 11.84KK136 pKa = 9.34KK137 pKa = 10.8
Molecular weight: 15.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2437 |
91 |
602 |
348.1 |
39.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.965 ± 0.258 | 2.216 ± 0.888 |
5.663 ± 0.371 | 6.689 ± 0.613 |
3.98 ± 0.377 | 5.827 ± 1.068 |
1.888 ± 0.205 | 4.924 ± 0.541 |
5.458 ± 0.922 | 9.315 ± 0.87 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.847 ± 0.415 | 5.006 ± 0.397 |
5.54 ± 0.968 | 3.939 ± 0.562 |
6.237 ± 0.804 | 8.289 ± 0.609 |
7.017 ± 1.209 | 6.648 ± 0.654 |
1.149 ± 0.312 | 3.406 ± 0.525 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
