
Pepper vein yellows virus 2
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Sobelivirales; Solemoviridae; Polerovirus
Average proteome isoelectric point is 7.87
Get precalculated fractions of proteins
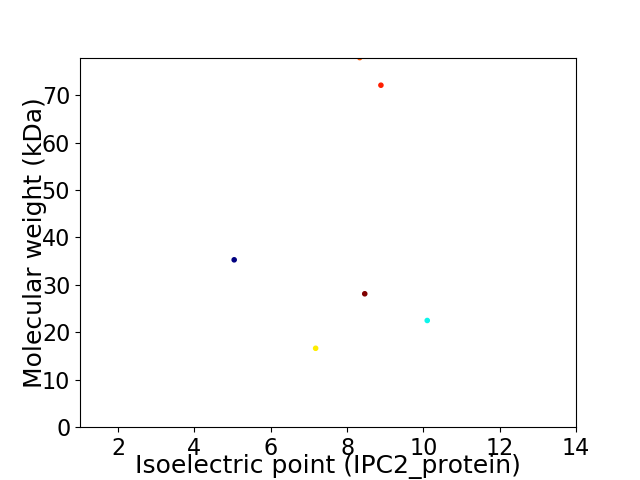
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|I3Y1T5|I3Y1T5_9LUTE Readthrough protein OS=Pepper vein yellows virus 2 OX=2560636 PE=3 SV=1
MM1 pKa = 7.85LEE3 pKa = 4.17VEE5 pKa = 4.53SSDD8 pKa = 3.37MSAEE12 pKa = 4.07EE13 pKa = 3.96LVQAGLCDD21 pKa = 4.92PIRR24 pKa = 11.84TFVKK28 pKa = 10.42RR29 pKa = 11.84EE30 pKa = 3.58PHH32 pKa = 5.86KK33 pKa = 10.47QSKK36 pKa = 10.11LDD38 pKa = 3.38EE39 pKa = 4.27GRR41 pKa = 11.84YY42 pKa = 8.96RR43 pKa = 11.84LIMSVSLVDD52 pKa = 3.45QLVARR57 pKa = 11.84VLFQNQNKK65 pKa = 9.66RR66 pKa = 11.84EE67 pKa = 3.96IALWRR72 pKa = 11.84ANPSKK77 pKa = 10.61PGFGLSTDD85 pKa = 3.63EE86 pKa = 4.0QVLEE90 pKa = 4.39FVQALAAQVEE100 pKa = 4.75VPPEE104 pKa = 3.73EE105 pKa = 5.59VITSWEE111 pKa = 4.24KK112 pKa = 10.58YY113 pKa = 9.87LVPTDD118 pKa = 3.67CSGFDD123 pKa = 3.21WSVAEE128 pKa = 4.57WMLHH132 pKa = 5.89DD133 pKa = 5.83DD134 pKa = 3.43MVVRR138 pKa = 11.84NKK140 pKa = 9.41LTLDD144 pKa = 4.03LNPTTEE150 pKa = 4.13KK151 pKa = 10.71LRR153 pKa = 11.84SVWLKK158 pKa = 9.68CICNSVLCLSDD169 pKa = 3.31GTLLAQRR176 pKa = 11.84VPGVQKK182 pKa = 10.45SGSYY186 pKa = 8.25NTSSSNSRR194 pKa = 11.84IRR196 pKa = 11.84VMAAYY201 pKa = 9.95HH202 pKa = 6.59CGADD206 pKa = 2.88WAMAMGDD213 pKa = 3.72DD214 pKa = 4.39ALEE217 pKa = 4.26SVNTNLEE224 pKa = 4.09VYY226 pKa = 10.15KK227 pKa = 10.94DD228 pKa = 3.51LGFKK232 pKa = 10.78VEE234 pKa = 4.14VSGQLEE240 pKa = 4.4FCSHH244 pKa = 6.9IFRR247 pKa = 11.84APDD250 pKa = 2.96LALPVNEE257 pKa = 4.57RR258 pKa = 11.84KK259 pKa = 8.59MLYY262 pKa = 10.45KK263 pKa = 10.76LIFGYY268 pKa = 10.86NPGSGNLEE276 pKa = 4.07VISNYY281 pKa = 9.46IAACVSVLNEE291 pKa = 4.24LRR293 pKa = 11.84HH294 pKa = 6.43DD295 pKa = 4.37PDD297 pKa = 3.73SVALLHH303 pKa = 5.94QWLVSPVLPQNNN315 pKa = 3.37
MM1 pKa = 7.85LEE3 pKa = 4.17VEE5 pKa = 4.53SSDD8 pKa = 3.37MSAEE12 pKa = 4.07EE13 pKa = 3.96LVQAGLCDD21 pKa = 4.92PIRR24 pKa = 11.84TFVKK28 pKa = 10.42RR29 pKa = 11.84EE30 pKa = 3.58PHH32 pKa = 5.86KK33 pKa = 10.47QSKK36 pKa = 10.11LDD38 pKa = 3.38EE39 pKa = 4.27GRR41 pKa = 11.84YY42 pKa = 8.96RR43 pKa = 11.84LIMSVSLVDD52 pKa = 3.45QLVARR57 pKa = 11.84VLFQNQNKK65 pKa = 9.66RR66 pKa = 11.84EE67 pKa = 3.96IALWRR72 pKa = 11.84ANPSKK77 pKa = 10.61PGFGLSTDD85 pKa = 3.63EE86 pKa = 4.0QVLEE90 pKa = 4.39FVQALAAQVEE100 pKa = 4.75VPPEE104 pKa = 3.73EE105 pKa = 5.59VITSWEE111 pKa = 4.24KK112 pKa = 10.58YY113 pKa = 9.87LVPTDD118 pKa = 3.67CSGFDD123 pKa = 3.21WSVAEE128 pKa = 4.57WMLHH132 pKa = 5.89DD133 pKa = 5.83DD134 pKa = 3.43MVVRR138 pKa = 11.84NKK140 pKa = 9.41LTLDD144 pKa = 4.03LNPTTEE150 pKa = 4.13KK151 pKa = 10.71LRR153 pKa = 11.84SVWLKK158 pKa = 9.68CICNSVLCLSDD169 pKa = 3.31GTLLAQRR176 pKa = 11.84VPGVQKK182 pKa = 10.45SGSYY186 pKa = 8.25NTSSSNSRR194 pKa = 11.84IRR196 pKa = 11.84VMAAYY201 pKa = 9.95HH202 pKa = 6.59CGADD206 pKa = 2.88WAMAMGDD213 pKa = 3.72DD214 pKa = 4.39ALEE217 pKa = 4.26SVNTNLEE224 pKa = 4.09VYY226 pKa = 10.15KK227 pKa = 10.94DD228 pKa = 3.51LGFKK232 pKa = 10.78VEE234 pKa = 4.14VSGQLEE240 pKa = 4.4FCSHH244 pKa = 6.9IFRR247 pKa = 11.84APDD250 pKa = 2.96LALPVNEE257 pKa = 4.57RR258 pKa = 11.84KK259 pKa = 8.59MLYY262 pKa = 10.45KK263 pKa = 10.76LIFGYY268 pKa = 10.86NPGSGNLEE276 pKa = 4.07VISNYY281 pKa = 9.46IAACVSVLNEE291 pKa = 4.24LRR293 pKa = 11.84HH294 pKa = 6.43DD295 pKa = 4.37PDD297 pKa = 3.73SVALLHH303 pKa = 5.94QWLVSPVLPQNNN315 pKa = 3.37
Molecular weight: 35.29 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E1A0W5|E1A0W5_9LUTE Movement protein OS=Pepper vein yellows virus 2 OX=2560636 PE=3 SV=2
MM1 pKa = 6.55NTEE4 pKa = 3.97GVRR7 pKa = 11.84RR8 pKa = 11.84NNNGNGGSRR17 pKa = 11.84GTRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84PRR26 pKa = 11.84QVRR29 pKa = 11.84PVVVVAPPGRR39 pKa = 11.84TRR41 pKa = 11.84RR42 pKa = 11.84GNRR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84SGGRR52 pKa = 11.84NRR54 pKa = 11.84NRR56 pKa = 11.84VGGRR60 pKa = 11.84SSNSEE65 pKa = 3.69TFIFNKK71 pKa = 10.42DD72 pKa = 3.59SIKK75 pKa = 10.74DD76 pKa = 3.69SSSGTVTFGPSLSEE90 pKa = 4.37SIALSGGVLKK100 pKa = 10.54AYY102 pKa = 10.22HH103 pKa = 6.68EE104 pKa = 4.5YY105 pKa = 10.51KK106 pKa = 9.32ITMVNIRR113 pKa = 11.84FVSEE117 pKa = 4.1SSSTAEE123 pKa = 3.6GSIAYY128 pKa = 9.39EE129 pKa = 4.08LDD131 pKa = 3.39PHH133 pKa = 7.02CKK135 pKa = 8.88LTSLQSTLRR144 pKa = 11.84KK145 pKa = 9.77FPVTKK150 pKa = 10.36GGQATFRR157 pKa = 11.84ASQINGVEE165 pKa = 3.87WHH167 pKa = 6.98DD168 pKa = 3.52TSEE171 pKa = 4.07DD172 pKa = 3.6QFRR175 pKa = 11.84LLYY178 pKa = 10.2KK179 pKa = 10.92GNGTKK184 pKa = 10.41GVAAGFFQIRR194 pKa = 11.84FTVQLHH200 pKa = 5.06NPKK203 pKa = 10.54
MM1 pKa = 6.55NTEE4 pKa = 3.97GVRR7 pKa = 11.84RR8 pKa = 11.84NNNGNGGSRR17 pKa = 11.84GTRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84PRR26 pKa = 11.84QVRR29 pKa = 11.84PVVVVAPPGRR39 pKa = 11.84TRR41 pKa = 11.84RR42 pKa = 11.84GNRR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84SGGRR52 pKa = 11.84NRR54 pKa = 11.84NRR56 pKa = 11.84VGGRR60 pKa = 11.84SSNSEE65 pKa = 3.69TFIFNKK71 pKa = 10.42DD72 pKa = 3.59SIKK75 pKa = 10.74DD76 pKa = 3.69SSSGTVTFGPSLSEE90 pKa = 4.37SIALSGGVLKK100 pKa = 10.54AYY102 pKa = 10.22HH103 pKa = 6.68EE104 pKa = 4.5YY105 pKa = 10.51KK106 pKa = 9.32ITMVNIRR113 pKa = 11.84FVSEE117 pKa = 4.1SSSTAEE123 pKa = 3.6GSIAYY128 pKa = 9.39EE129 pKa = 4.08LDD131 pKa = 3.39PHH133 pKa = 7.02CKK135 pKa = 8.88LTSLQSTLRR144 pKa = 11.84KK145 pKa = 9.77FPVTKK150 pKa = 10.36GGQATFRR157 pKa = 11.84ASQINGVEE165 pKa = 3.87WHH167 pKa = 6.98DD168 pKa = 3.52TSEE171 pKa = 4.07DD172 pKa = 3.6QFRR175 pKa = 11.84LLYY178 pKa = 10.2KK179 pKa = 10.92GNGTKK184 pKa = 10.41GVAAGFFQIRR194 pKa = 11.84FTVQLHH200 pKa = 5.06NPKK203 pKa = 10.54
Molecular weight: 22.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2273 |
153 |
699 |
378.8 |
42.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.379 ± 0.72 | 1.452 ± 0.364 |
4.179 ± 0.916 | 6.115 ± 0.277 |
3.784 ± 0.397 | 7.479 ± 0.695 |
2.156 ± 0.444 | 3.696 ± 0.23 |
6.071 ± 0.694 | 8.447 ± 1.284 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.716 ± 0.279 | 4.663 ± 0.635 |
6.467 ± 0.752 | 3.784 ± 0.158 |
6.775 ± 0.979 | 9.855 ± 0.548 |
6.027 ± 0.739 | 6.467 ± 0.704 |
1.804 ± 0.192 | 2.64 ± 0.33 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
