
Micromonas commoda (strain RCC299 / NOUM17 / CCMP2709) (Picoplanktonic green alga)
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; Mamiellophyceae; Mamiellales; Mamiellaceae; Micromonas
Average proteome isoelectric point is 6.5
Get precalculated fractions of proteins
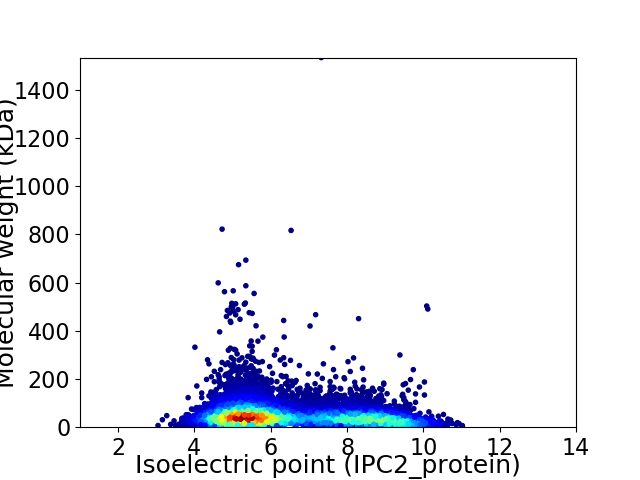
Virtual 2D-PAGE plot for 10115 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
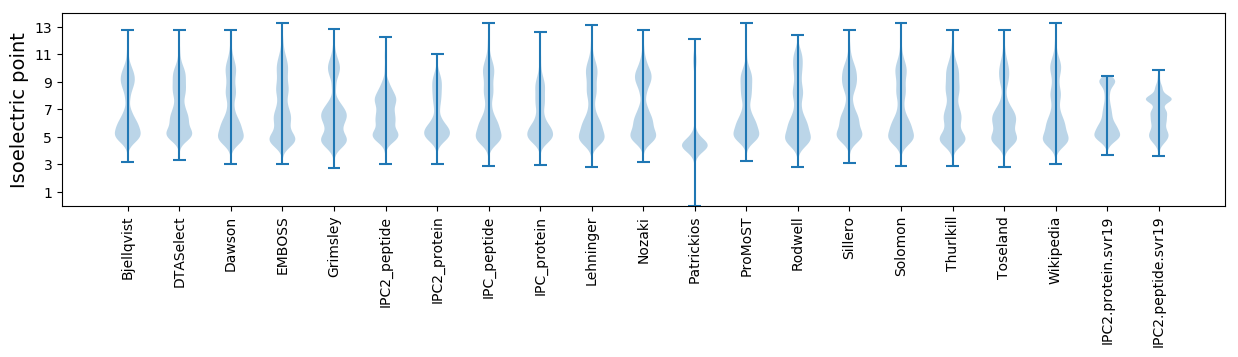
Protein with the lowest isoelectric point:
>tr|C1ECW3|C1ECW3_MICCC Fe2OG dioxygenase domain-containing protein OS=Micromonas commoda (strain RCC299 / NOUM17 / CCMP2709) OX=296587 GN=MICPUN_61562 PE=4 SV=1
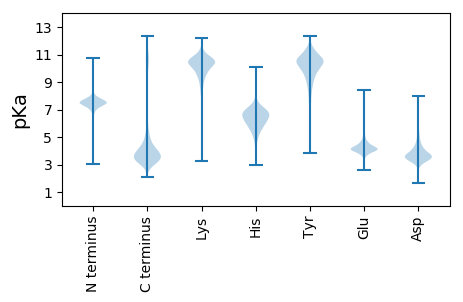
MM1 pKa = 7.51VIGAYY6 pKa = 10.35GDD8 pKa = 3.83DD9 pKa = 4.55DD10 pKa = 4.48KK11 pKa = 12.05GSDD14 pKa = 3.24SGLAYY19 pKa = 10.82VFTRR23 pKa = 11.84NTAGDD28 pKa = 4.02LASGWTQVAKK38 pKa = 9.54LTASDD43 pKa = 4.24GAAGDD48 pKa = 3.55QFGQSVSIDD57 pKa = 3.2GDD59 pKa = 4.19TVVIGAWRR67 pKa = 11.84DD68 pKa = 3.4DD69 pKa = 4.01DD70 pKa = 4.55KK71 pKa = 11.82GSEE74 pKa = 4.09SGSAYY79 pKa = 10.45VFTRR83 pKa = 11.84VTAGDD88 pKa = 4.11LASDD92 pKa = 3.95WTQVAKK98 pKa = 9.54LTASDD103 pKa = 3.88GAGYY107 pKa = 10.47NYY109 pKa = 10.35FGISVSIDD117 pKa = 3.1GDD119 pKa = 4.01TMVIGANYY127 pKa = 10.35DD128 pKa = 3.42DD129 pKa = 5.55DD130 pKa = 5.41NGIEE134 pKa = 4.14SGSAYY139 pKa = 10.66VFTRR143 pKa = 11.84NTAGDD148 pKa = 4.02LASGWTQVAKK158 pKa = 9.54LTASDD163 pKa = 3.77GAHH166 pKa = 6.35YY167 pKa = 11.01DD168 pKa = 3.32NFGYY172 pKa = 10.12SVSIDD177 pKa = 3.19GDD179 pKa = 4.14TVVIGARR186 pKa = 11.84YY187 pKa = 10.25DD188 pKa = 3.55DD189 pKa = 4.96DD190 pKa = 4.87KK191 pKa = 11.79GSNSGSAYY199 pKa = 10.29VFTRR203 pKa = 11.84NTAGDD208 pKa = 4.02LASGWTQVAKK218 pKa = 9.46LTSSDD223 pKa = 2.92GAANDD228 pKa = 3.43RR229 pKa = 11.84FGRR232 pKa = 11.84SVSIDD237 pKa = 3.04GDD239 pKa = 4.19TVVIGAYY246 pKa = 10.29ADD248 pKa = 4.93DD249 pKa = 6.09DD250 pKa = 4.98DD251 pKa = 4.82GTDD254 pKa = 3.13SGSAYY259 pKa = 10.75VFTRR263 pKa = 11.84DD264 pKa = 3.23TAGDD268 pKa = 3.9LASGWTQVAKK278 pKa = 9.55LTADD282 pKa = 3.97DD283 pKa = 4.98GAAGDD288 pKa = 3.73NFGWSVSIDD297 pKa = 3.21GDD299 pKa = 4.16TVVIGSQRR307 pKa = 11.84DD308 pKa = 3.12ADD310 pKa = 3.86NGIEE314 pKa = 4.1SGSAYY319 pKa = 10.7VFTRR323 pKa = 11.84DD324 pKa = 3.23TAGDD328 pKa = 3.92LASSWTQFAKK338 pKa = 9.57LTAYY342 pKa = 10.41DD343 pKa = 3.82GAGYY347 pKa = 10.22DD348 pKa = 3.57YY349 pKa = 10.71FGWSVSIDD357 pKa = 3.54GDD359 pKa = 4.0TMVIGAYY366 pKa = 9.6RR367 pKa = 11.84DD368 pKa = 3.34SHH370 pKa = 6.21IANDD374 pKa = 3.35VGSAYY379 pKa = 10.56VFEE382 pKa = 4.96FALTPGSCSVSAPPRR397 pKa = 11.84NGNGNDD403 pKa = 3.61CVFPLAHH410 pKa = 7.03GEE412 pKa = 4.06ACTPVCDD419 pKa = 4.26HH420 pKa = 6.74GFTAVTASAGCVNGNLFPGEE440 pKa = 4.35CRR442 pKa = 11.84CSRR445 pKa = 11.84GYY447 pKa = 9.44PRR449 pKa = 11.84SYY451 pKa = 10.08HH452 pKa = 6.4GNWPCSIEE460 pKa = 4.51KK461 pKa = 10.49RR462 pKa = 11.84MEE464 pKa = 3.83LL465 pKa = 3.92
MM1 pKa = 7.51VIGAYY6 pKa = 10.35GDD8 pKa = 3.83DD9 pKa = 4.55DD10 pKa = 4.48KK11 pKa = 12.05GSDD14 pKa = 3.24SGLAYY19 pKa = 10.82VFTRR23 pKa = 11.84NTAGDD28 pKa = 4.02LASGWTQVAKK38 pKa = 9.54LTASDD43 pKa = 4.24GAAGDD48 pKa = 3.55QFGQSVSIDD57 pKa = 3.2GDD59 pKa = 4.19TVVIGAWRR67 pKa = 11.84DD68 pKa = 3.4DD69 pKa = 4.01DD70 pKa = 4.55KK71 pKa = 11.82GSEE74 pKa = 4.09SGSAYY79 pKa = 10.45VFTRR83 pKa = 11.84VTAGDD88 pKa = 4.11LASDD92 pKa = 3.95WTQVAKK98 pKa = 9.54LTASDD103 pKa = 3.88GAGYY107 pKa = 10.47NYY109 pKa = 10.35FGISVSIDD117 pKa = 3.1GDD119 pKa = 4.01TMVIGANYY127 pKa = 10.35DD128 pKa = 3.42DD129 pKa = 5.55DD130 pKa = 5.41NGIEE134 pKa = 4.14SGSAYY139 pKa = 10.66VFTRR143 pKa = 11.84NTAGDD148 pKa = 4.02LASGWTQVAKK158 pKa = 9.54LTASDD163 pKa = 3.77GAHH166 pKa = 6.35YY167 pKa = 11.01DD168 pKa = 3.32NFGYY172 pKa = 10.12SVSIDD177 pKa = 3.19GDD179 pKa = 4.14TVVIGARR186 pKa = 11.84YY187 pKa = 10.25DD188 pKa = 3.55DD189 pKa = 4.96DD190 pKa = 4.87KK191 pKa = 11.79GSNSGSAYY199 pKa = 10.29VFTRR203 pKa = 11.84NTAGDD208 pKa = 4.02LASGWTQVAKK218 pKa = 9.46LTSSDD223 pKa = 2.92GAANDD228 pKa = 3.43RR229 pKa = 11.84FGRR232 pKa = 11.84SVSIDD237 pKa = 3.04GDD239 pKa = 4.19TVVIGAYY246 pKa = 10.29ADD248 pKa = 4.93DD249 pKa = 6.09DD250 pKa = 4.98DD251 pKa = 4.82GTDD254 pKa = 3.13SGSAYY259 pKa = 10.75VFTRR263 pKa = 11.84DD264 pKa = 3.23TAGDD268 pKa = 3.9LASGWTQVAKK278 pKa = 9.55LTADD282 pKa = 3.97DD283 pKa = 4.98GAAGDD288 pKa = 3.73NFGWSVSIDD297 pKa = 3.21GDD299 pKa = 4.16TVVIGSQRR307 pKa = 11.84DD308 pKa = 3.12ADD310 pKa = 3.86NGIEE314 pKa = 4.1SGSAYY319 pKa = 10.7VFTRR323 pKa = 11.84DD324 pKa = 3.23TAGDD328 pKa = 3.92LASSWTQFAKK338 pKa = 9.57LTAYY342 pKa = 10.41DD343 pKa = 3.82GAGYY347 pKa = 10.22DD348 pKa = 3.57YY349 pKa = 10.71FGWSVSIDD357 pKa = 3.54GDD359 pKa = 4.0TMVIGAYY366 pKa = 9.6RR367 pKa = 11.84DD368 pKa = 3.34SHH370 pKa = 6.21IANDD374 pKa = 3.35VGSAYY379 pKa = 10.56VFEE382 pKa = 4.96FALTPGSCSVSAPPRR397 pKa = 11.84NGNGNDD403 pKa = 3.61CVFPLAHH410 pKa = 7.03GEE412 pKa = 4.06ACTPVCDD419 pKa = 4.26HH420 pKa = 6.74GFTAVTASAGCVNGNLFPGEE440 pKa = 4.35CRR442 pKa = 11.84CSRR445 pKa = 11.84GYY447 pKa = 9.44PRR449 pKa = 11.84SYY451 pKa = 10.08HH452 pKa = 6.4GNWPCSIEE460 pKa = 4.51KK461 pKa = 10.49RR462 pKa = 11.84MEE464 pKa = 3.83LL465 pKa = 3.92
Molecular weight: 48.26 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C1E8X9|C1E8X9_MICCC Patatin OS=Micromonas commoda (strain RCC299 / NOUM17 / CCMP2709) OX=296587 GN=MICPUN_59530 PE=3 SV=1
MM1 pKa = 7.75APPAPPGGWGRR12 pKa = 11.84PVSAPAGTGPPSRR25 pKa = 11.84VGPPGGRR32 pKa = 11.84VGPPGGRR39 pKa = 11.84VGPPGGAPARR49 pKa = 11.84FGPSRR54 pKa = 11.84VGPGRR59 pKa = 11.84QQ60 pKa = 2.98
MM1 pKa = 7.75APPAPPGGWGRR12 pKa = 11.84PVSAPAGTGPPSRR25 pKa = 11.84VGPPGGRR32 pKa = 11.84VGPPGGRR39 pKa = 11.84VGPPGGAPARR49 pKa = 11.84FGPSRR54 pKa = 11.84VGPGRR59 pKa = 11.84QQ60 pKa = 2.98
Molecular weight: 5.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
4889369 |
24 |
14149 |
483.4 |
52.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.618 ± 0.054 | 1.574 ± 0.013 |
6.465 ± 0.023 | 6.747 ± 0.032 |
3.147 ± 0.017 | 8.819 ± 0.044 |
2.06 ± 0.011 | 3.192 ± 0.022 |
4.441 ± 0.026 | 7.986 ± 0.03 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.197 ± 0.013 | 2.71 ± 0.013 |
5.56 ± 0.028 | 2.579 ± 0.015 |
7.656 ± 0.038 | 6.571 ± 0.028 |
5.13 ± 0.019 | 7.222 ± 0.022 |
1.303 ± 0.009 | 2.021 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
