
Oceanicaulis sp. HTCC2633
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Maricaulales; Maricaulaceae; Oceanicaulis; unclassified Oceanicaulis
Average proteome isoelectric point is 5.8
Get precalculated fractions of proteins
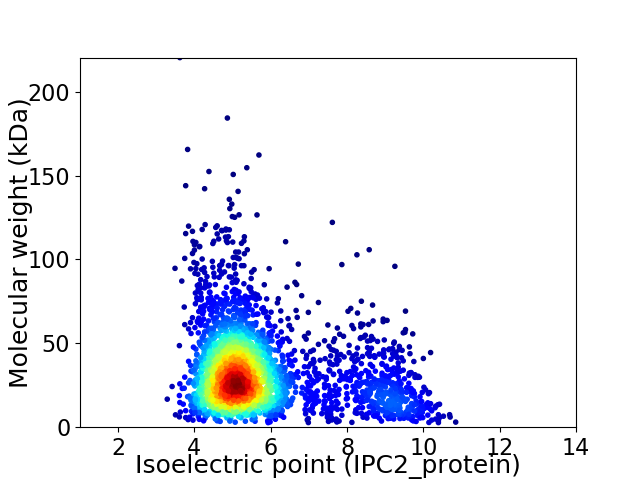
Virtual 2D-PAGE plot for 3028 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A3UEP8|A3UEP8_9PROT Membrane protein putative OS=Oceanicaulis sp. HTCC2633 OX=314254 GN=OA2633_11660 PE=4 SV=1
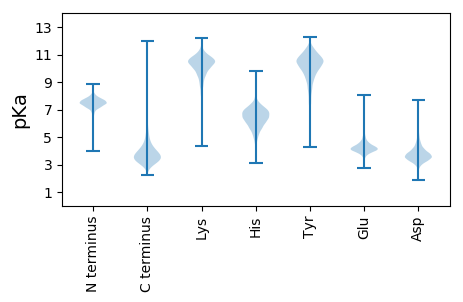
MM1 pKa = 7.06TSKK4 pKa = 11.16LILSVKK10 pKa = 10.25GLVLFAWLSLFSATALAQDD29 pKa = 4.75LGSSQSIDD37 pKa = 3.28FNAAGFPDD45 pKa = 4.25PALLLGKK52 pKa = 9.34PYY54 pKa = 10.75ANSGFTFDD62 pKa = 4.46YY63 pKa = 10.96SGNLYY68 pKa = 10.95GLDD71 pKa = 3.66EE72 pKa = 4.73SDD74 pKa = 3.95ARR76 pKa = 11.84QGKK79 pKa = 9.01LWVQSATQDD88 pKa = 3.2EE89 pKa = 5.49TFTIDD94 pKa = 3.39YY95 pKa = 10.92SGDD98 pKa = 3.63NFDD101 pKa = 4.97LNSFILVNGFSTYY114 pKa = 10.66NITVRR119 pKa = 11.84GYY121 pKa = 9.45DD122 pKa = 3.09ASNSVVEE129 pKa = 4.42TQSFSVPSGTQTISLTNSGFNNVRR153 pKa = 11.84YY154 pKa = 10.06VIITATSANGFDD166 pKa = 4.49GSFFDD171 pKa = 5.38DD172 pKa = 4.2FAVAPPTVPDD182 pKa = 3.32VTAPRR187 pKa = 11.84VTSIVRR193 pKa = 11.84QTPATAITSVDD204 pKa = 3.38SLVFRR209 pKa = 11.84VTFDD213 pKa = 3.36EE214 pKa = 4.5AVQNVSSADD223 pKa = 3.51FTVTGVSGAALGVSGSSAVYY243 pKa = 10.39DD244 pKa = 3.46ITVSGGDD251 pKa = 3.34TASYY255 pKa = 9.58TGTVSLGFRR264 pKa = 11.84GTQDD268 pKa = 3.27IADD271 pKa = 4.13TPAGNTLVNTTPTGANEE288 pKa = 4.93SYY290 pKa = 10.16TLDD293 pKa = 3.51NTAPAAPSTPDD304 pKa = 3.5LDD306 pKa = 4.63AGSDD310 pKa = 3.56TGSSNTDD317 pKa = 3.15NITNDD322 pKa = 3.38TTPTFTGTAEE332 pKa = 4.02ANATVTVSSTLSGPLGSTTADD353 pKa = 3.3GSGNWSYY360 pKa = 11.53TSGVLAEE367 pKa = 4.45GAHH370 pKa = 6.54TITATATDD378 pKa = 3.48AASNVSAASSALSITIDD395 pKa = 3.23TTAPDD400 pKa = 3.17ITSITRR406 pKa = 11.84LSPTGQTISASAANAAFRR424 pKa = 11.84VTYY427 pKa = 10.38SEE429 pKa = 5.21AIAGSSQADD438 pKa = 3.4YY439 pKa = 11.51AFQNVSGTAAGSVSSYY455 pKa = 9.06GTSSPTLINVQLGTLSGDD473 pKa = 3.27GDD475 pKa = 4.11FSITLNGGHH484 pKa = 7.69GITDD488 pKa = 3.32IAGNALSSTTPTGVRR503 pKa = 11.84EE504 pKa = 3.99VFTRR508 pKa = 11.84DD509 pKa = 2.64GTAPSTTSFVRR520 pKa = 11.84TTPAGATTSSDD531 pKa = 3.43SLVFTANYY539 pKa = 9.01SEE541 pKa = 4.58SVSGVDD547 pKa = 3.14TSDD550 pKa = 3.58FAVTGTTATVQSVSAASGTSIAITVSGGDD579 pKa = 3.28LASYY583 pKa = 10.6NGTVGLNYY591 pKa = 10.18AAAPTITDD599 pKa = 3.39AAGNALPNTEE609 pKa = 4.15PATDD613 pKa = 3.39EE614 pKa = 4.7TYY616 pKa = 10.8TLDD619 pKa = 3.35NTAPAGYY626 pKa = 9.95SVTLDD631 pKa = 4.21DD632 pKa = 5.86DD633 pKa = 4.03PVNAGNQTATSFTFAGAEE651 pKa = 3.86VAANYY656 pKa = 10.46NYY658 pKa = 9.9TISSSGGGANVTGSGTISTATDD680 pKa = 3.44QISGIDD686 pKa = 3.59VSGLGDD692 pKa = 3.55GTLTLSVTLTDD703 pKa = 3.87GAGNAGSAATDD714 pKa = 3.54TATKK718 pKa = 9.71EE719 pKa = 4.05AQPPQVNSIVVAGSPAGNATSVDD742 pKa = 4.01FTVTFSEE749 pKa = 4.3AAQNISTDD757 pKa = 3.94DD758 pKa = 3.78FSLTTTGTATGTIASVSSATGTTVTVTVNTISGDD792 pKa = 3.36GTIRR796 pKa = 11.84LDD798 pKa = 4.33LDD800 pKa = 3.65AATNIQDD807 pKa = 3.54AVGNSGPNAYY817 pKa = 9.16TSGATHH823 pKa = 6.31TVDD826 pKa = 3.12NTAPAAFNVAFQAGLITQANEE847 pKa = 3.6TSNDD851 pKa = 2.81IRR853 pKa = 11.84FTTAEE858 pKa = 3.94TGTTYY863 pKa = 10.69AYY865 pKa = 10.4SISSSGGGTPVTGTGTITTATQDD888 pKa = 3.12VTVNVSSLPDD898 pKa = 3.38GTLTLTATLTDD909 pKa = 3.55GAGNVTNQSSADD921 pKa = 3.81TVSKK925 pKa = 9.79ATAVPGFSMAFAPTSVLTGGTSTATT950 pKa = 3.33
MM1 pKa = 7.06TSKK4 pKa = 11.16LILSVKK10 pKa = 10.25GLVLFAWLSLFSATALAQDD29 pKa = 4.75LGSSQSIDD37 pKa = 3.28FNAAGFPDD45 pKa = 4.25PALLLGKK52 pKa = 9.34PYY54 pKa = 10.75ANSGFTFDD62 pKa = 4.46YY63 pKa = 10.96SGNLYY68 pKa = 10.95GLDD71 pKa = 3.66EE72 pKa = 4.73SDD74 pKa = 3.95ARR76 pKa = 11.84QGKK79 pKa = 9.01LWVQSATQDD88 pKa = 3.2EE89 pKa = 5.49TFTIDD94 pKa = 3.39YY95 pKa = 10.92SGDD98 pKa = 3.63NFDD101 pKa = 4.97LNSFILVNGFSTYY114 pKa = 10.66NITVRR119 pKa = 11.84GYY121 pKa = 9.45DD122 pKa = 3.09ASNSVVEE129 pKa = 4.42TQSFSVPSGTQTISLTNSGFNNVRR153 pKa = 11.84YY154 pKa = 10.06VIITATSANGFDD166 pKa = 4.49GSFFDD171 pKa = 5.38DD172 pKa = 4.2FAVAPPTVPDD182 pKa = 3.32VTAPRR187 pKa = 11.84VTSIVRR193 pKa = 11.84QTPATAITSVDD204 pKa = 3.38SLVFRR209 pKa = 11.84VTFDD213 pKa = 3.36EE214 pKa = 4.5AVQNVSSADD223 pKa = 3.51FTVTGVSGAALGVSGSSAVYY243 pKa = 10.39DD244 pKa = 3.46ITVSGGDD251 pKa = 3.34TASYY255 pKa = 9.58TGTVSLGFRR264 pKa = 11.84GTQDD268 pKa = 3.27IADD271 pKa = 4.13TPAGNTLVNTTPTGANEE288 pKa = 4.93SYY290 pKa = 10.16TLDD293 pKa = 3.51NTAPAAPSTPDD304 pKa = 3.5LDD306 pKa = 4.63AGSDD310 pKa = 3.56TGSSNTDD317 pKa = 3.15NITNDD322 pKa = 3.38TTPTFTGTAEE332 pKa = 4.02ANATVTVSSTLSGPLGSTTADD353 pKa = 3.3GSGNWSYY360 pKa = 11.53TSGVLAEE367 pKa = 4.45GAHH370 pKa = 6.54TITATATDD378 pKa = 3.48AASNVSAASSALSITIDD395 pKa = 3.23TTAPDD400 pKa = 3.17ITSITRR406 pKa = 11.84LSPTGQTISASAANAAFRR424 pKa = 11.84VTYY427 pKa = 10.38SEE429 pKa = 5.21AIAGSSQADD438 pKa = 3.4YY439 pKa = 11.51AFQNVSGTAAGSVSSYY455 pKa = 9.06GTSSPTLINVQLGTLSGDD473 pKa = 3.27GDD475 pKa = 4.11FSITLNGGHH484 pKa = 7.69GITDD488 pKa = 3.32IAGNALSSTTPTGVRR503 pKa = 11.84EE504 pKa = 3.99VFTRR508 pKa = 11.84DD509 pKa = 2.64GTAPSTTSFVRR520 pKa = 11.84TTPAGATTSSDD531 pKa = 3.43SLVFTANYY539 pKa = 9.01SEE541 pKa = 4.58SVSGVDD547 pKa = 3.14TSDD550 pKa = 3.58FAVTGTTATVQSVSAASGTSIAITVSGGDD579 pKa = 3.28LASYY583 pKa = 10.6NGTVGLNYY591 pKa = 10.18AAAPTITDD599 pKa = 3.39AAGNALPNTEE609 pKa = 4.15PATDD613 pKa = 3.39EE614 pKa = 4.7TYY616 pKa = 10.8TLDD619 pKa = 3.35NTAPAGYY626 pKa = 9.95SVTLDD631 pKa = 4.21DD632 pKa = 5.86DD633 pKa = 4.03PVNAGNQTATSFTFAGAEE651 pKa = 3.86VAANYY656 pKa = 10.46NYY658 pKa = 9.9TISSSGGGANVTGSGTISTATDD680 pKa = 3.44QISGIDD686 pKa = 3.59VSGLGDD692 pKa = 3.55GTLTLSVTLTDD703 pKa = 3.87GAGNAGSAATDD714 pKa = 3.54TATKK718 pKa = 9.71EE719 pKa = 4.05AQPPQVNSIVVAGSPAGNATSVDD742 pKa = 4.01FTVTFSEE749 pKa = 4.3AAQNISTDD757 pKa = 3.94DD758 pKa = 3.78FSLTTTGTATGTIASVSSATGTTVTVTVNTISGDD792 pKa = 3.36GTIRR796 pKa = 11.84LDD798 pKa = 4.33LDD800 pKa = 3.65AATNIQDD807 pKa = 3.54AVGNSGPNAYY817 pKa = 9.16TSGATHH823 pKa = 6.31TVDD826 pKa = 3.12NTAPAAFNVAFQAGLITQANEE847 pKa = 3.6TSNDD851 pKa = 2.81IRR853 pKa = 11.84FTTAEE858 pKa = 3.94TGTTYY863 pKa = 10.69AYY865 pKa = 10.4SISSSGGGTPVTGTGTITTATQDD888 pKa = 3.12VTVNVSSLPDD898 pKa = 3.38GTLTLTATLTDD909 pKa = 3.55GAGNVTNQSSADD921 pKa = 3.81TVSKK925 pKa = 9.79ATAVPGFSMAFAPTSVLTGGTSTATT950 pKa = 3.33
Molecular weight: 94.72 kDa
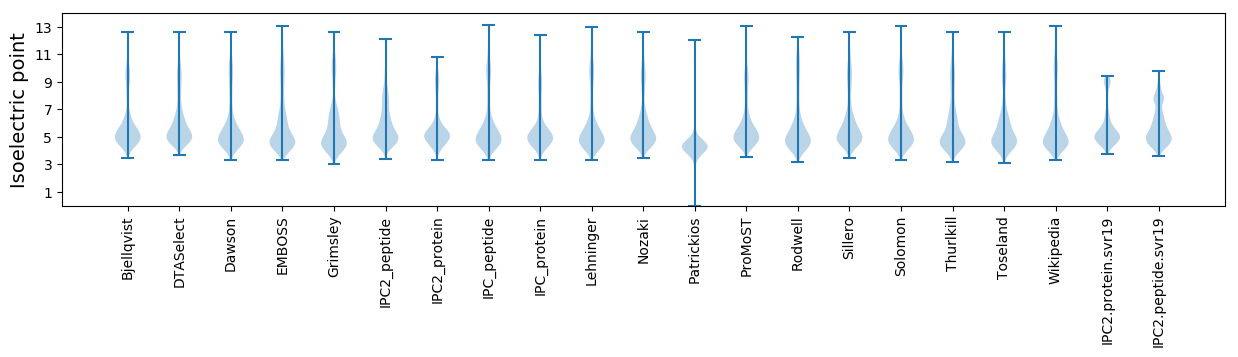
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A3UC48|A3UC48_9PROT Uncharacterized protein OS=Oceanicaulis sp. HTCC2633 OX=314254 GN=OA2633_01606 PE=4 SV=1
SS1 pKa = 7.65PMRR4 pKa = 11.84QNMKK8 pKa = 9.59IRR10 pKa = 11.84RR11 pKa = 11.84AMPMVAVVRR20 pKa = 11.84KK21 pKa = 9.27AQARR25 pKa = 11.84LL26 pKa = 3.28
SS1 pKa = 7.65PMRR4 pKa = 11.84QNMKK8 pKa = 9.59IRR10 pKa = 11.84RR11 pKa = 11.84AMPMVAVVRR20 pKa = 11.84KK21 pKa = 9.27AQARR25 pKa = 11.84LL26 pKa = 3.28
Molecular weight: 3.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
953775 |
23 |
2187 |
315.0 |
34.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.869 ± 0.059 | 0.746 ± 0.014 |
6.38 ± 0.036 | 6.581 ± 0.05 |
3.67 ± 0.028 | 8.505 ± 0.04 |
1.872 ± 0.027 | 4.705 ± 0.026 |
2.832 ± 0.038 | 10.161 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.412 ± 0.023 | 2.551 ± 0.03 |
4.958 ± 0.033 | 3.323 ± 0.027 |
7.004 ± 0.042 | 5.643 ± 0.035 |
5.114 ± 0.039 | 7.14 ± 0.033 |
1.318 ± 0.018 | 2.215 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
