
Ahrensia marina
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Ahrensiaceae; Ahrensia
Average proteome isoelectric point is 6.17
Get precalculated fractions of proteins
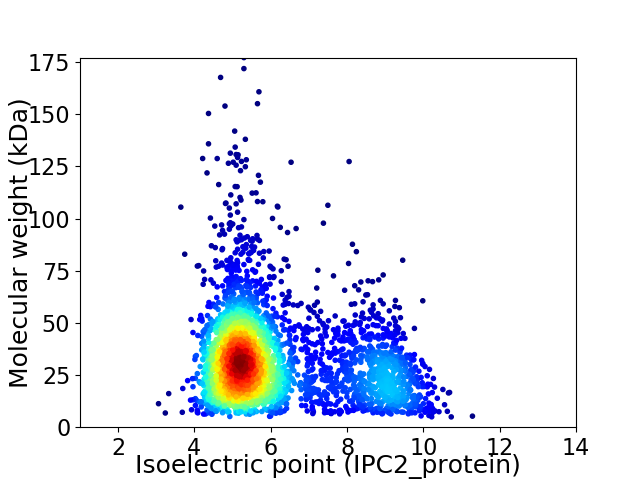
Virtual 2D-PAGE plot for 3161 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0M9GNV3|A0A0M9GNV3_9RHOB Nitroreductase OS=Ahrensia marina OX=1514904 GN=SU32_04965 PE=4 SV=1
TT1 pKa = 7.62IEE3 pKa = 5.19DD4 pKa = 3.63GGAVSNGDD12 pKa = 3.45GVIGNDD18 pKa = 3.2SDD20 pKa = 4.37GVGAVTVTGSGSTWDD35 pKa = 3.42NTSTLRR41 pKa = 11.84VGNAGTGTLTIADD54 pKa = 4.14GGAVSVNSGTGTVMIADD71 pKa = 4.34DD72 pKa = 4.68AGSAGALNIGDD83 pKa = 4.03GASAGILNAATVDD96 pKa = 3.91GGSGAATLNFKK107 pKa = 8.79HH108 pKa = 6.55TDD110 pKa = 2.67TDD112 pKa = 4.12YY113 pKa = 11.35FFTNDD118 pKa = 3.13GTSAGAAIDD127 pKa = 3.37ITGTVAVNHH136 pKa = 6.34TATGTTTLTGTNTYY150 pKa = 9.2TGSTIVNFGTLAVDD164 pKa = 4.41GGSIAHH170 pKa = 6.64TSADD174 pKa = 4.15LIVGGFSSDD183 pKa = 4.92DD184 pKa = 3.31GTLTIKK190 pKa = 10.93NGGSVSNNEE199 pKa = 3.69AGIGYY204 pKa = 9.51AAGSEE209 pKa = 4.58GEE211 pKa = 4.29VSVTGAGSNWTNSGTLYY228 pKa = 10.81VGLAGSGTVTIADD241 pKa = 3.97GGSVSNTAGTVGSASGAIGTVTVTGAGSSWSSSSDD276 pKa = 3.41LYY278 pKa = 11.25VGEE281 pKa = 4.82SGTGTMNVEE290 pKa = 4.59DD291 pKa = 4.5GATAFVGDD299 pKa = 4.21DD300 pKa = 3.38VVLGFNATGNGTLTVDD316 pKa = 3.7SVGSEE321 pKa = 4.0LDD323 pKa = 2.98ITNAFVVGRR332 pKa = 11.84AGIGALKK339 pKa = 9.21VQNGGTLTGNYY350 pKa = 9.58SYY352 pKa = 11.43VGNSSGATGTATITGSGSSWSLTGDD377 pKa = 4.2LYY379 pKa = 10.65IAEE382 pKa = 4.43QGTGTMTIADD392 pKa = 4.32GGTVSNAVSYY402 pKa = 10.48IGYY405 pKa = 10.3DD406 pKa = 3.3SGSDD410 pKa = 3.45GAVTVTGAGSTWTNNSTLYY429 pKa = 10.76VGEE432 pKa = 5.16DD433 pKa = 3.59GNGTLTISDD442 pKa = 4.28GGTVSAVGGVEE453 pKa = 3.75IASASGSSVINIGAAEE469 pKa = 4.14GNAAVAAGTLDD480 pKa = 3.03TSAIVFGSGNGEE492 pKa = 3.84LVFNHH497 pKa = 6.87TDD499 pKa = 2.9TAYY502 pKa = 10.74DD503 pKa = 3.62FDD505 pKa = 4.18AAIFGLGSIEE515 pKa = 4.3HH516 pKa = 6.29FAGVTNLTADD526 pKa = 3.22NSGFTGEE533 pKa = 4.52TVVSGGSLYY542 pKa = 11.37VNDD545 pKa = 4.79SIGGTVYY552 pKa = 10.89VDD554 pKa = 4.31DD555 pKa = 4.56GTLGGSGTTGALTVNFGGNLAPGNSIGTLNVASAIFNAGSTYY597 pKa = 9.21TVEE600 pKa = 4.14LNNGGFVAGTNNDD613 pKa = 3.73LLNATGVVTINGGTVNVTSEE633 pKa = 4.0NGTEE637 pKa = 4.23DD638 pKa = 3.42GSTYY642 pKa = 9.46ATPGTYY648 pKa = 9.84TIITGGSVTGTFDD661 pKa = 3.71NVSDD665 pKa = 4.79DD666 pKa = 3.97YY667 pKa = 12.1VFLDD671 pKa = 3.98FTDD674 pKa = 4.83SYY676 pKa = 11.86DD677 pKa = 3.38ATNVYY682 pKa = 8.31LTSEE686 pKa = 4.12QVVFFSDD693 pKa = 3.06IAEE696 pKa = 4.42TPNQQAIASPLEE708 pKa = 3.93ALGSGNTVYY717 pKa = 10.82DD718 pKa = 3.81ALVGLVGDD726 pKa = 4.41EE727 pKa = 4.28DD728 pKa = 3.99DD729 pKa = 4.64ARR731 pKa = 11.84AAFDD735 pKa = 3.65SLTGEE740 pKa = 4.87IYY742 pKa = 10.83ASAQTALLEE751 pKa = 4.29DD752 pKa = 3.32SRR754 pKa = 11.84LPRR757 pKa = 11.84EE758 pKa = 3.95AAMEE762 pKa = 4.9RR763 pKa = 11.84IRR765 pKa = 11.84HH766 pKa = 5.38AFDD769 pKa = 5.94GIGTDD774 pKa = 4.07NSAQTEE780 pKa = 4.13DD781 pKa = 4.53RR782 pKa = 11.84ISEE785 pKa = 4.32SFGLWSQGFGAWSQWKK801 pKa = 10.1SDD803 pKa = 3.64GNAATMDD810 pKa = 3.7RR811 pKa = 11.84SIGGLLVGGDD821 pKa = 3.58TMASDD826 pKa = 3.71NVRR829 pKa = 11.84FGMLGGYY836 pKa = 8.95SRR838 pKa = 11.84SHH840 pKa = 6.63LNLDD844 pKa = 3.36NRR846 pKa = 11.84LSSGTAEE853 pKa = 4.24TYY855 pKa = 10.04TLGVYY860 pKa = 10.43GGGEE864 pKa = 3.62WDD866 pKa = 3.66AFSLKK871 pKa = 10.69GGIAHH876 pKa = 6.74SWHH879 pKa = 6.35NLDD882 pKa = 3.34TSRR885 pKa = 11.84SVAFNGFSDD894 pKa = 4.05ILSASYY900 pKa = 10.55NARR903 pKa = 11.84TLQAWGEE910 pKa = 4.12LAVSFDD916 pKa = 3.33TDD918 pKa = 2.96IARR921 pKa = 11.84FEE923 pKa = 4.11PFANLTHH930 pKa = 6.37VNLNIDD936 pKa = 3.84GFTEE940 pKa = 4.11TGGAAALTAAPNNTDD955 pKa = 2.92ATFLTVGLRR964 pKa = 11.84AEE966 pKa = 4.55TEE968 pKa = 4.22VSLGSADD975 pKa = 3.24VTLRR979 pKa = 11.84GTVAWQHH986 pKa = 5.92AFSNVPTSQMSFASGDD1002 pKa = 3.7SPFTIDD1008 pKa = 5.4GVPLAQDD1015 pKa = 3.62SLALGAGFDD1024 pKa = 4.19VNLTDD1029 pKa = 4.37SAKK1032 pKa = 10.79LDD1034 pKa = 3.69FSYY1037 pKa = 11.05DD1038 pKa = 3.12GRR1040 pKa = 11.84FGSDD1044 pKa = 3.09VHH1046 pKa = 6.72DD1047 pKa = 4.67HH1048 pKa = 6.26SATVSLNVLFF1058 pKa = 5.17
TT1 pKa = 7.62IEE3 pKa = 5.19DD4 pKa = 3.63GGAVSNGDD12 pKa = 3.45GVIGNDD18 pKa = 3.2SDD20 pKa = 4.37GVGAVTVTGSGSTWDD35 pKa = 3.42NTSTLRR41 pKa = 11.84VGNAGTGTLTIADD54 pKa = 4.14GGAVSVNSGTGTVMIADD71 pKa = 4.34DD72 pKa = 4.68AGSAGALNIGDD83 pKa = 4.03GASAGILNAATVDD96 pKa = 3.91GGSGAATLNFKK107 pKa = 8.79HH108 pKa = 6.55TDD110 pKa = 2.67TDD112 pKa = 4.12YY113 pKa = 11.35FFTNDD118 pKa = 3.13GTSAGAAIDD127 pKa = 3.37ITGTVAVNHH136 pKa = 6.34TATGTTTLTGTNTYY150 pKa = 9.2TGSTIVNFGTLAVDD164 pKa = 4.41GGSIAHH170 pKa = 6.64TSADD174 pKa = 4.15LIVGGFSSDD183 pKa = 4.92DD184 pKa = 3.31GTLTIKK190 pKa = 10.93NGGSVSNNEE199 pKa = 3.69AGIGYY204 pKa = 9.51AAGSEE209 pKa = 4.58GEE211 pKa = 4.29VSVTGAGSNWTNSGTLYY228 pKa = 10.81VGLAGSGTVTIADD241 pKa = 3.97GGSVSNTAGTVGSASGAIGTVTVTGAGSSWSSSSDD276 pKa = 3.41LYY278 pKa = 11.25VGEE281 pKa = 4.82SGTGTMNVEE290 pKa = 4.59DD291 pKa = 4.5GATAFVGDD299 pKa = 4.21DD300 pKa = 3.38VVLGFNATGNGTLTVDD316 pKa = 3.7SVGSEE321 pKa = 4.0LDD323 pKa = 2.98ITNAFVVGRR332 pKa = 11.84AGIGALKK339 pKa = 9.21VQNGGTLTGNYY350 pKa = 9.58SYY352 pKa = 11.43VGNSSGATGTATITGSGSSWSLTGDD377 pKa = 4.2LYY379 pKa = 10.65IAEE382 pKa = 4.43QGTGTMTIADD392 pKa = 4.32GGTVSNAVSYY402 pKa = 10.48IGYY405 pKa = 10.3DD406 pKa = 3.3SGSDD410 pKa = 3.45GAVTVTGAGSTWTNNSTLYY429 pKa = 10.76VGEE432 pKa = 5.16DD433 pKa = 3.59GNGTLTISDD442 pKa = 4.28GGTVSAVGGVEE453 pKa = 3.75IASASGSSVINIGAAEE469 pKa = 4.14GNAAVAAGTLDD480 pKa = 3.03TSAIVFGSGNGEE492 pKa = 3.84LVFNHH497 pKa = 6.87TDD499 pKa = 2.9TAYY502 pKa = 10.74DD503 pKa = 3.62FDD505 pKa = 4.18AAIFGLGSIEE515 pKa = 4.3HH516 pKa = 6.29FAGVTNLTADD526 pKa = 3.22NSGFTGEE533 pKa = 4.52TVVSGGSLYY542 pKa = 11.37VNDD545 pKa = 4.79SIGGTVYY552 pKa = 10.89VDD554 pKa = 4.31DD555 pKa = 4.56GTLGGSGTTGALTVNFGGNLAPGNSIGTLNVASAIFNAGSTYY597 pKa = 9.21TVEE600 pKa = 4.14LNNGGFVAGTNNDD613 pKa = 3.73LLNATGVVTINGGTVNVTSEE633 pKa = 4.0NGTEE637 pKa = 4.23DD638 pKa = 3.42GSTYY642 pKa = 9.46ATPGTYY648 pKa = 9.84TIITGGSVTGTFDD661 pKa = 3.71NVSDD665 pKa = 4.79DD666 pKa = 3.97YY667 pKa = 12.1VFLDD671 pKa = 3.98FTDD674 pKa = 4.83SYY676 pKa = 11.86DD677 pKa = 3.38ATNVYY682 pKa = 8.31LTSEE686 pKa = 4.12QVVFFSDD693 pKa = 3.06IAEE696 pKa = 4.42TPNQQAIASPLEE708 pKa = 3.93ALGSGNTVYY717 pKa = 10.82DD718 pKa = 3.81ALVGLVGDD726 pKa = 4.41EE727 pKa = 4.28DD728 pKa = 3.99DD729 pKa = 4.64ARR731 pKa = 11.84AAFDD735 pKa = 3.65SLTGEE740 pKa = 4.87IYY742 pKa = 10.83ASAQTALLEE751 pKa = 4.29DD752 pKa = 3.32SRR754 pKa = 11.84LPRR757 pKa = 11.84EE758 pKa = 3.95AAMEE762 pKa = 4.9RR763 pKa = 11.84IRR765 pKa = 11.84HH766 pKa = 5.38AFDD769 pKa = 5.94GIGTDD774 pKa = 4.07NSAQTEE780 pKa = 4.13DD781 pKa = 4.53RR782 pKa = 11.84ISEE785 pKa = 4.32SFGLWSQGFGAWSQWKK801 pKa = 10.1SDD803 pKa = 3.64GNAATMDD810 pKa = 3.7RR811 pKa = 11.84SIGGLLVGGDD821 pKa = 3.58TMASDD826 pKa = 3.71NVRR829 pKa = 11.84FGMLGGYY836 pKa = 8.95SRR838 pKa = 11.84SHH840 pKa = 6.63LNLDD844 pKa = 3.36NRR846 pKa = 11.84LSSGTAEE853 pKa = 4.24TYY855 pKa = 10.04TLGVYY860 pKa = 10.43GGGEE864 pKa = 3.62WDD866 pKa = 3.66AFSLKK871 pKa = 10.69GGIAHH876 pKa = 6.74SWHH879 pKa = 6.35NLDD882 pKa = 3.34TSRR885 pKa = 11.84SVAFNGFSDD894 pKa = 4.05ILSASYY900 pKa = 10.55NARR903 pKa = 11.84TLQAWGEE910 pKa = 4.12LAVSFDD916 pKa = 3.33TDD918 pKa = 2.96IARR921 pKa = 11.84FEE923 pKa = 4.11PFANLTHH930 pKa = 6.37VNLNIDD936 pKa = 3.84GFTEE940 pKa = 4.11TGGAAALTAAPNNTDD955 pKa = 2.92ATFLTVGLRR964 pKa = 11.84AEE966 pKa = 4.55TEE968 pKa = 4.22VSLGSADD975 pKa = 3.24VTLRR979 pKa = 11.84GTVAWQHH986 pKa = 5.92AFSNVPTSQMSFASGDD1002 pKa = 3.7SPFTIDD1008 pKa = 5.4GVPLAQDD1015 pKa = 3.62SLALGAGFDD1024 pKa = 4.19VNLTDD1029 pKa = 4.37SAKK1032 pKa = 10.79LDD1034 pKa = 3.69FSYY1037 pKa = 11.05DD1038 pKa = 3.12GRR1040 pKa = 11.84FGSDD1044 pKa = 3.09VHH1046 pKa = 6.72DD1047 pKa = 4.67HH1048 pKa = 6.26SATVSLNVLFF1058 pKa = 5.17
Molecular weight: 105.48 kDa
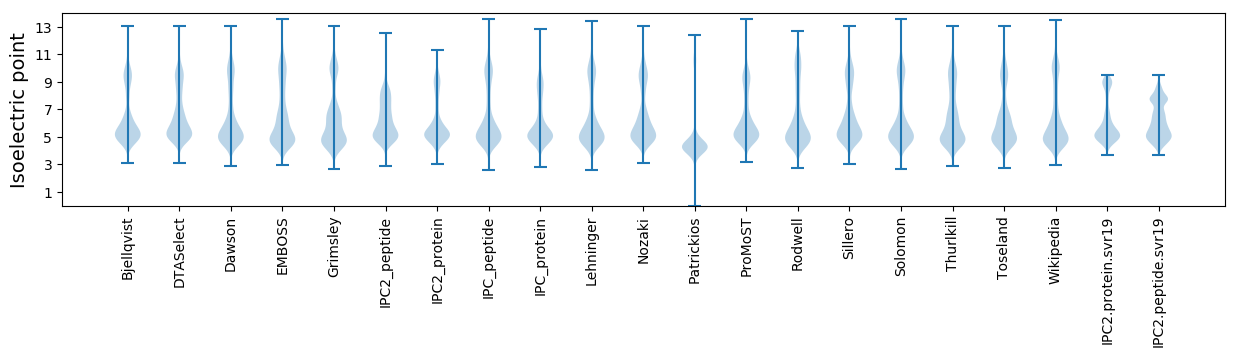
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0M9GLI9|A0A0M9GLI9_9RHOB HAD family hydrolase OS=Ahrensia marina OX=1514904 GN=SU32_12980 PE=4 SV=1
MM1 pKa = 6.23STKK4 pKa = 9.65RR5 pKa = 11.84TFQPSRR11 pKa = 11.84LVRR14 pKa = 11.84ARR16 pKa = 11.84RR17 pKa = 11.84HH18 pKa = 4.67GFRR21 pKa = 11.84SRR23 pKa = 11.84MATAAGRR30 pKa = 11.84NVLARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84AKK39 pKa = 10.17GRR41 pKa = 11.84KK42 pKa = 8.79KK43 pKa = 10.63LSAA46 pKa = 3.95
MM1 pKa = 6.23STKK4 pKa = 9.65RR5 pKa = 11.84TFQPSRR11 pKa = 11.84LVRR14 pKa = 11.84ARR16 pKa = 11.84RR17 pKa = 11.84HH18 pKa = 4.67GFRR21 pKa = 11.84SRR23 pKa = 11.84MATAAGRR30 pKa = 11.84NVLARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84AKK39 pKa = 10.17GRR41 pKa = 11.84KK42 pKa = 8.79KK43 pKa = 10.63LSAA46 pKa = 3.95
Molecular weight: 5.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
971630 |
41 |
1601 |
307.4 |
33.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.74 ± 0.049 | 0.886 ± 0.014 |
5.952 ± 0.036 | 6.173 ± 0.039 |
4.129 ± 0.033 | 7.982 ± 0.04 |
1.98 ± 0.024 | 6.376 ± 0.035 |
4.73 ± 0.035 | 9.489 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.833 ± 0.022 | 3.565 ± 0.023 |
4.381 ± 0.026 | 3.268 ± 0.025 |
5.499 ± 0.042 | 5.994 ± 0.03 |
5.377 ± 0.026 | 6.992 ± 0.031 |
1.213 ± 0.019 | 2.442 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
