
Denitrovibrio acetiphilus (strain DSM 12809 / NBRC 114555 / N2460)
Taxonomy: cellular organisms; Bacteria; Deferribacteres; Deferribacteres; Deferribacterales; Deferribacteraceae; Denitrovibrio; Denitrovibrio acetiphilus
Average proteome isoelectric point is 6.25
Get precalculated fractions of proteins
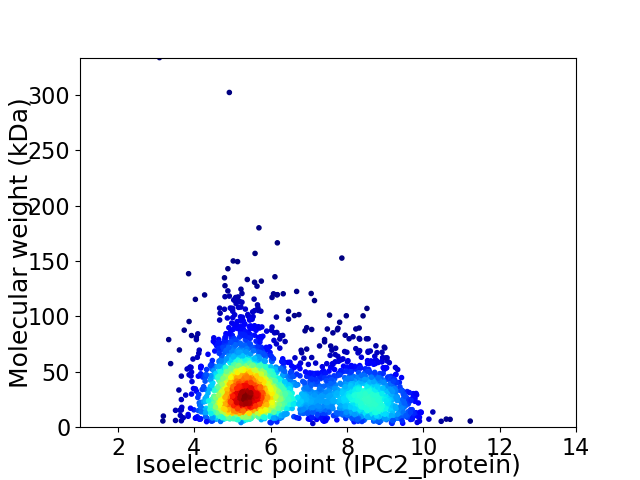
Virtual 2D-PAGE plot for 2901 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D4H2R2|D4H2R2_DENA2 DUF374 domain-containing protein OS=Denitrovibrio acetiphilus (strain DSM 12809 / NBRC 114555 / N2460) OX=522772 GN=Dacet_0323 PE=4 SV=1
MM1 pKa = 6.99MRR3 pKa = 11.84SLYY6 pKa = 10.63SAISGLSVNQQAMDD20 pKa = 3.5VLGNNISNVNTVGYY34 pKa = 10.08KK35 pKa = 9.53SGRR38 pKa = 11.84AVFEE42 pKa = 4.8DD43 pKa = 4.09MLSQTLVGSTAPSDD57 pKa = 3.69TLGGTNSTQVGLGTSLAGVDD77 pKa = 4.13TIFEE81 pKa = 4.39TGSMQSTSVNTDD93 pKa = 3.27LAIQGDD99 pKa = 4.42GFFVIKK105 pKa = 10.47GAGSSDD111 pKa = 3.56LLYY114 pKa = 10.73TRR116 pKa = 11.84AGDD119 pKa = 3.95FRR121 pKa = 11.84FDD123 pKa = 3.4TSGSLVTSSGFYY135 pKa = 8.23VQGWMVDD142 pKa = 3.34PDD144 pKa = 3.73TGEE147 pKa = 4.4LLTEE151 pKa = 4.63GTTEE155 pKa = 5.23DD156 pKa = 3.45IVLSSAYY163 pKa = 8.17QTAQAKK169 pKa = 6.81VTSEE173 pKa = 3.69ASLAGVLDD181 pKa = 3.84TDD183 pKa = 4.46ADD185 pKa = 4.11PSIVSYY191 pKa = 10.63PSLLHH196 pKa = 6.15YY197 pKa = 10.82ADD199 pKa = 4.08TGDD202 pKa = 5.44SIFQVYY208 pKa = 10.53SDD210 pKa = 5.19DD211 pKa = 3.97GANMDD216 pKa = 4.25LADD219 pKa = 4.09NEE221 pKa = 4.72SIKK224 pKa = 11.04VKK226 pKa = 10.7AHH228 pKa = 4.75GTGMTEE234 pKa = 3.11MGYY237 pKa = 9.69IYY239 pKa = 9.63NTSDD243 pKa = 3.24VNLDD247 pKa = 3.86LASDD251 pKa = 3.97PQLLVYY257 pKa = 10.56INSNPVTLEE266 pKa = 3.6YY267 pKa = 10.9GVDD270 pKa = 3.94FNTMAEE276 pKa = 4.16LANALEE282 pKa = 4.46TQLNTAAGAADD293 pKa = 4.14FSVAVVDD300 pKa = 3.7GTINVTRR307 pKa = 11.84NTVVGANVSIDD318 pKa = 3.8SFSGSPLLAVAMSDD332 pKa = 3.45LASVYY337 pKa = 11.09DD338 pKa = 3.98DD339 pKa = 5.12AGDD342 pKa = 3.77TKK344 pKa = 11.16SSDD347 pKa = 2.83EE348 pKa = 4.48MYY350 pKa = 10.81YY351 pKa = 10.54EE352 pKa = 4.08SQIYY356 pKa = 9.95AGRR359 pKa = 11.84DD360 pKa = 3.43FTTLTEE366 pKa = 3.93LASAIEE372 pKa = 3.96EE373 pKa = 4.68AIDD376 pKa = 3.88GNVIPTDD383 pKa = 3.94EE384 pKa = 4.53FNVTYY389 pKa = 10.71DD390 pKa = 3.13SATGQFNYY398 pKa = 10.55VIDD401 pKa = 3.86NTLNSALGTLRR412 pKa = 11.84LSGFSLDD419 pKa = 3.4KK420 pKa = 10.71AYY422 pKa = 11.14SGTVFEE428 pKa = 4.92NNIVPAGAVIEE439 pKa = 4.31AVYY442 pKa = 8.78GTTDD446 pKa = 3.37TEE448 pKa = 4.33LSNTFLRR455 pKa = 11.84TAQDD459 pKa = 3.9DD460 pKa = 4.17DD461 pKa = 4.38PLTEE465 pKa = 5.0LFTSSGDD472 pKa = 3.93SLGLDD477 pKa = 3.28ATAILQFSAGVAGEE491 pKa = 4.14NLAGTATIPAATSTLDD507 pKa = 3.66DD508 pKa = 3.99LRR510 pKa = 11.84EE511 pKa = 4.27AIAAYY516 pKa = 9.04MGYY519 pKa = 10.83DD520 pKa = 3.25SATEE524 pKa = 4.02DD525 pKa = 5.2DD526 pKa = 4.51VLQHH530 pKa = 6.54IASLDD535 pKa = 3.74KK536 pKa = 11.45NSGKK540 pKa = 10.12LQITGEE546 pKa = 4.14KK547 pKa = 10.27GSPNEE552 pKa = 3.38IDD554 pKa = 3.5FVKK557 pKa = 10.91FEE559 pKa = 4.44VVGSGTYY566 pKa = 9.0DD567 pKa = 3.07TFYY570 pKa = 11.09DD571 pKa = 3.9YY572 pKa = 11.81YY573 pKa = 10.79EE574 pKa = 4.55YY575 pKa = 10.6NTVQEE580 pKa = 4.52AKK582 pKa = 10.46GGTLTTSQTIYY593 pKa = 10.49DD594 pKa = 4.01AQGNAHH600 pKa = 5.13TVQYY604 pKa = 9.86NFEE607 pKa = 4.36MSDD610 pKa = 3.37SLDD613 pKa = 3.87NVWKK617 pKa = 9.21LTVSTPDD624 pKa = 3.47DD625 pKa = 3.66GAEE628 pKa = 3.59ISFNEE633 pKa = 4.1TTGNQVYY640 pKa = 7.3MHH642 pKa = 6.68FNANGSFNYY651 pKa = 8.53LTTEE655 pKa = 3.73SGQRR659 pKa = 11.84IASLSMNYY667 pKa = 8.41DD668 pKa = 3.13TGGGAGVVSDD678 pKa = 4.44IEE680 pKa = 4.26MDD682 pKa = 3.47LGTAGKK688 pKa = 10.16FDD690 pKa = 5.12GIYY693 pKa = 10.12ISSDD697 pKa = 2.93DD698 pKa = 4.21SGITKK703 pKa = 10.16AAQDD707 pKa = 4.12GYY709 pKa = 10.92ATGLLEE715 pKa = 4.19STLFNSNGEE724 pKa = 4.02IIGYY728 pKa = 6.03YY729 pKa = 10.2TNGQVATIAQIATATFTNPGGLTKK753 pKa = 10.76VGDD756 pKa = 3.62TTFQKK761 pKa = 10.67SSNSGDD767 pKa = 3.63PAIGRR772 pKa = 11.84PGTGFRR778 pKa = 11.84GKK780 pKa = 9.87VAAEE784 pKa = 3.75TLEE787 pKa = 4.0NSNVDD792 pKa = 3.51LSTEE796 pKa = 4.12FVNMITTQRR805 pKa = 11.84GFQANSRR812 pKa = 11.84VITTSDD818 pKa = 3.03EE819 pKa = 4.06MLQEE823 pKa = 4.04LMSLKK828 pKa = 10.43RR829 pKa = 3.65
MM1 pKa = 6.99MRR3 pKa = 11.84SLYY6 pKa = 10.63SAISGLSVNQQAMDD20 pKa = 3.5VLGNNISNVNTVGYY34 pKa = 10.08KK35 pKa = 9.53SGRR38 pKa = 11.84AVFEE42 pKa = 4.8DD43 pKa = 4.09MLSQTLVGSTAPSDD57 pKa = 3.69TLGGTNSTQVGLGTSLAGVDD77 pKa = 4.13TIFEE81 pKa = 4.39TGSMQSTSVNTDD93 pKa = 3.27LAIQGDD99 pKa = 4.42GFFVIKK105 pKa = 10.47GAGSSDD111 pKa = 3.56LLYY114 pKa = 10.73TRR116 pKa = 11.84AGDD119 pKa = 3.95FRR121 pKa = 11.84FDD123 pKa = 3.4TSGSLVTSSGFYY135 pKa = 8.23VQGWMVDD142 pKa = 3.34PDD144 pKa = 3.73TGEE147 pKa = 4.4LLTEE151 pKa = 4.63GTTEE155 pKa = 5.23DD156 pKa = 3.45IVLSSAYY163 pKa = 8.17QTAQAKK169 pKa = 6.81VTSEE173 pKa = 3.69ASLAGVLDD181 pKa = 3.84TDD183 pKa = 4.46ADD185 pKa = 4.11PSIVSYY191 pKa = 10.63PSLLHH196 pKa = 6.15YY197 pKa = 10.82ADD199 pKa = 4.08TGDD202 pKa = 5.44SIFQVYY208 pKa = 10.53SDD210 pKa = 5.19DD211 pKa = 3.97GANMDD216 pKa = 4.25LADD219 pKa = 4.09NEE221 pKa = 4.72SIKK224 pKa = 11.04VKK226 pKa = 10.7AHH228 pKa = 4.75GTGMTEE234 pKa = 3.11MGYY237 pKa = 9.69IYY239 pKa = 9.63NTSDD243 pKa = 3.24VNLDD247 pKa = 3.86LASDD251 pKa = 3.97PQLLVYY257 pKa = 10.56INSNPVTLEE266 pKa = 3.6YY267 pKa = 10.9GVDD270 pKa = 3.94FNTMAEE276 pKa = 4.16LANALEE282 pKa = 4.46TQLNTAAGAADD293 pKa = 4.14FSVAVVDD300 pKa = 3.7GTINVTRR307 pKa = 11.84NTVVGANVSIDD318 pKa = 3.8SFSGSPLLAVAMSDD332 pKa = 3.45LASVYY337 pKa = 11.09DD338 pKa = 3.98DD339 pKa = 5.12AGDD342 pKa = 3.77TKK344 pKa = 11.16SSDD347 pKa = 2.83EE348 pKa = 4.48MYY350 pKa = 10.81YY351 pKa = 10.54EE352 pKa = 4.08SQIYY356 pKa = 9.95AGRR359 pKa = 11.84DD360 pKa = 3.43FTTLTEE366 pKa = 3.93LASAIEE372 pKa = 3.96EE373 pKa = 4.68AIDD376 pKa = 3.88GNVIPTDD383 pKa = 3.94EE384 pKa = 4.53FNVTYY389 pKa = 10.71DD390 pKa = 3.13SATGQFNYY398 pKa = 10.55VIDD401 pKa = 3.86NTLNSALGTLRR412 pKa = 11.84LSGFSLDD419 pKa = 3.4KK420 pKa = 10.71AYY422 pKa = 11.14SGTVFEE428 pKa = 4.92NNIVPAGAVIEE439 pKa = 4.31AVYY442 pKa = 8.78GTTDD446 pKa = 3.37TEE448 pKa = 4.33LSNTFLRR455 pKa = 11.84TAQDD459 pKa = 3.9DD460 pKa = 4.17DD461 pKa = 4.38PLTEE465 pKa = 5.0LFTSSGDD472 pKa = 3.93SLGLDD477 pKa = 3.28ATAILQFSAGVAGEE491 pKa = 4.14NLAGTATIPAATSTLDD507 pKa = 3.66DD508 pKa = 3.99LRR510 pKa = 11.84EE511 pKa = 4.27AIAAYY516 pKa = 9.04MGYY519 pKa = 10.83DD520 pKa = 3.25SATEE524 pKa = 4.02DD525 pKa = 5.2DD526 pKa = 4.51VLQHH530 pKa = 6.54IASLDD535 pKa = 3.74KK536 pKa = 11.45NSGKK540 pKa = 10.12LQITGEE546 pKa = 4.14KK547 pKa = 10.27GSPNEE552 pKa = 3.38IDD554 pKa = 3.5FVKK557 pKa = 10.91FEE559 pKa = 4.44VVGSGTYY566 pKa = 9.0DD567 pKa = 3.07TFYY570 pKa = 11.09DD571 pKa = 3.9YY572 pKa = 11.81YY573 pKa = 10.79EE574 pKa = 4.55YY575 pKa = 10.6NTVQEE580 pKa = 4.52AKK582 pKa = 10.46GGTLTTSQTIYY593 pKa = 10.49DD594 pKa = 4.01AQGNAHH600 pKa = 5.13TVQYY604 pKa = 9.86NFEE607 pKa = 4.36MSDD610 pKa = 3.37SLDD613 pKa = 3.87NVWKK617 pKa = 9.21LTVSTPDD624 pKa = 3.47DD625 pKa = 3.66GAEE628 pKa = 3.59ISFNEE633 pKa = 4.1TTGNQVYY640 pKa = 7.3MHH642 pKa = 6.68FNANGSFNYY651 pKa = 8.53LTTEE655 pKa = 3.73SGQRR659 pKa = 11.84IASLSMNYY667 pKa = 8.41DD668 pKa = 3.13TGGGAGVVSDD678 pKa = 4.44IEE680 pKa = 4.26MDD682 pKa = 3.47LGTAGKK688 pKa = 10.16FDD690 pKa = 5.12GIYY693 pKa = 10.12ISSDD697 pKa = 2.93DD698 pKa = 4.21SGITKK703 pKa = 10.16AAQDD707 pKa = 4.12GYY709 pKa = 10.92ATGLLEE715 pKa = 4.19STLFNSNGEE724 pKa = 4.02IIGYY728 pKa = 6.03YY729 pKa = 10.2TNGQVATIAQIATATFTNPGGLTKK753 pKa = 10.76VGDD756 pKa = 3.62TTFQKK761 pKa = 10.67SSNSGDD767 pKa = 3.63PAIGRR772 pKa = 11.84PGTGFRR778 pKa = 11.84GKK780 pKa = 9.87VAAEE784 pKa = 3.75TLEE787 pKa = 4.0NSNVDD792 pKa = 3.51LSTEE796 pKa = 4.12FVNMITTQRR805 pKa = 11.84GFQANSRR812 pKa = 11.84VITTSDD818 pKa = 3.03EE819 pKa = 4.06MLQEE823 pKa = 4.04LMSLKK828 pKa = 10.43RR829 pKa = 3.65
Molecular weight: 87.6 kDa
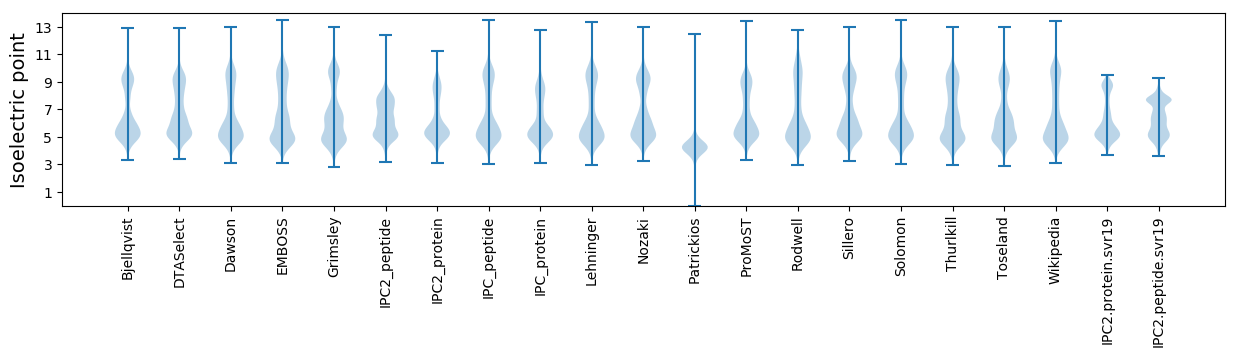
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D4H199|D4H199_DENA2 Short-chain dehydrogenase/reductase SDR OS=Denitrovibrio acetiphilus (strain DSM 12809 / NBRC 114555 / N2460) OX=522772 GN=Dacet_0041 PE=4 SV=1
MM1 pKa = 7.38AQMLTMKK8 pKa = 10.31KK9 pKa = 10.04PSNTKK14 pKa = 9.75RR15 pKa = 11.84KK16 pKa = 8.87RR17 pKa = 11.84KK18 pKa = 9.05HH19 pKa = 4.73GFRR22 pKa = 11.84ARR24 pKa = 11.84MKK26 pKa = 9.83TRR28 pKa = 11.84GGKK31 pKa = 9.28LAIARR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84AKK40 pKa = 9.74GRR42 pKa = 11.84KK43 pKa = 8.88RR44 pKa = 11.84LTVV47 pKa = 3.11
MM1 pKa = 7.38AQMLTMKK8 pKa = 10.31KK9 pKa = 10.04PSNTKK14 pKa = 9.75RR15 pKa = 11.84KK16 pKa = 8.87RR17 pKa = 11.84KK18 pKa = 9.05HH19 pKa = 4.73GFRR22 pKa = 11.84ARR24 pKa = 11.84MKK26 pKa = 9.83TRR28 pKa = 11.84GGKK31 pKa = 9.28LAIARR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84AKK40 pKa = 9.74GRR42 pKa = 11.84KK43 pKa = 8.88RR44 pKa = 11.84LTVV47 pKa = 3.11
Molecular weight: 5.51 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
946645 |
32 |
3226 |
326.3 |
36.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.345 ± 0.044 | 1.208 ± 0.019 |
6.024 ± 0.051 | 6.95 ± 0.047 |
4.643 ± 0.035 | 6.894 ± 0.042 |
1.834 ± 0.022 | 7.607 ± 0.042 |
7.203 ± 0.051 | 8.931 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.028 ± 0.022 | 4.568 ± 0.037 |
3.342 ± 0.029 | 2.649 ± 0.024 |
4.218 ± 0.033 | 6.598 ± 0.04 |
5.362 ± 0.044 | 7.049 ± 0.048 |
0.746 ± 0.015 | 3.801 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
