
Litorivita pollutaquae
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Litorivita
Average proteome isoelectric point is 6.15
Get precalculated fractions of proteins
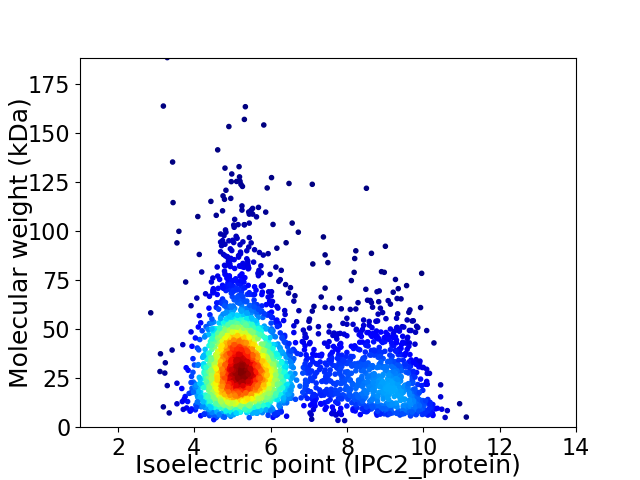
Virtual 2D-PAGE plot for 3171 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2V4MUX0|A0A2V4MUX0_9RHOB Uncharacterized protein OS=Litorivita pollutaquae OX=2200892 GN=DI396_08480 PE=4 SV=1
MM1 pKa = 8.12DD2 pKa = 4.77AQDD5 pKa = 3.51SAGNQATQRR14 pKa = 11.84SFVVTVTDD22 pKa = 3.62TTPPNAPVIGTMGATPDD39 pKa = 3.57SRR41 pKa = 11.84VGLEE45 pKa = 4.04GTAEE49 pKa = 4.33PGSAVAITFPDD60 pKa = 4.06GSTQSVIADD69 pKa = 3.58AGTGAFTVTSNTPQQSGVVTLVATDD94 pKa = 3.76AEE96 pKa = 5.01GNDD99 pKa = 4.14SVPTTAGFTGDD110 pKa = 4.14DD111 pKa = 3.91TPPTILIGALSGPTSGTYY129 pKa = 9.37IAAITLSEE137 pKa = 4.72DD138 pKa = 3.33STDD141 pKa = 4.5FDD143 pKa = 5.04AADD146 pKa = 3.69LTLTNATATLTGSGSDD162 pKa = 3.54YY163 pKa = 10.81TATLTPAADD172 pKa = 3.84GTVSLSVAAGVFTDD186 pKa = 4.59AAGNSNGASNTVSAVFAGVAPTVSISGAPEE216 pKa = 3.68ALAGGSSFTVAIAFSEE232 pKa = 4.68VVTGFVAADD241 pKa = 3.61ISATNATVTALTGGGVSYY259 pKa = 10.92VATVHH264 pKa = 6.95ASGTGDD270 pKa = 3.23VRR272 pKa = 11.84VAVPANVAFGATGHH286 pKa = 6.49GNLASSQVVIADD298 pKa = 3.22VSVQQTQEE306 pKa = 4.22HH307 pKa = 6.42IASYY311 pKa = 8.62MQTRR315 pKa = 11.84ANQLISNQPSLMSFLSGEE333 pKa = 4.06PRR335 pKa = 11.84GKK337 pKa = 10.77FNFAVTRR344 pKa = 11.84GAGSFDD350 pKa = 3.57VATGATGARR359 pKa = 11.84YY360 pKa = 8.2PVWAQANGSWTNDD373 pKa = 2.62GDD375 pKa = 4.23SEE377 pKa = 4.58SEE379 pKa = 4.46YY380 pKa = 11.33VFGALGGHH388 pKa = 4.73QTISEE393 pKa = 4.13NLLVGAMLQFDD404 pKa = 4.68HH405 pKa = 7.2LSEE408 pKa = 5.47DD409 pKa = 3.43EE410 pKa = 4.41GVASVRR416 pKa = 11.84GTGWMVGPYY425 pKa = 9.53FVARR429 pKa = 11.84SATQPLYY436 pKa = 10.91FEE438 pKa = 5.18GRR440 pKa = 11.84LLYY443 pKa = 11.05GEE445 pKa = 4.48TSNKK449 pKa = 9.32ISPFGTYY456 pKa = 9.63EE457 pKa = 4.1DD458 pKa = 4.53SFDD461 pKa = 3.74TTRR464 pKa = 11.84LLAQLKK470 pKa = 9.6VAGEE474 pKa = 4.08LSYY477 pKa = 11.44GRR479 pKa = 11.84TLLTPFMDD487 pKa = 4.11ASYY490 pKa = 9.26STDD493 pKa = 3.47DD494 pKa = 3.1QHH496 pKa = 9.94SYY498 pKa = 11.18VDD500 pKa = 3.75SLGNTIPGQDD510 pKa = 2.95IALGQIEE517 pKa = 4.42IGMDD521 pKa = 3.97FSMMLPVTTGDD532 pKa = 3.41LEE534 pKa = 4.27LWGGISGIWSHH545 pKa = 7.35IDD547 pKa = 3.08GSGYY551 pKa = 10.95ASTVTPDD558 pKa = 3.44YY559 pKa = 10.22EE560 pKa = 4.22GGRR563 pKa = 11.84ARR565 pKa = 11.84VEE567 pKa = 3.83LGINHH572 pKa = 6.26QDD574 pKa = 3.43SAGHH578 pKa = 6.2NFTVGTYY585 pKa = 10.24YY586 pKa = 10.9DD587 pKa = 4.23GIGANGYY594 pKa = 9.24EE595 pKa = 4.38SYY597 pKa = 11.06GLSLGYY603 pKa = 10.59EE604 pKa = 4.11MQFF607 pKa = 3.35
MM1 pKa = 8.12DD2 pKa = 4.77AQDD5 pKa = 3.51SAGNQATQRR14 pKa = 11.84SFVVTVTDD22 pKa = 3.62TTPPNAPVIGTMGATPDD39 pKa = 3.57SRR41 pKa = 11.84VGLEE45 pKa = 4.04GTAEE49 pKa = 4.33PGSAVAITFPDD60 pKa = 4.06GSTQSVIADD69 pKa = 3.58AGTGAFTVTSNTPQQSGVVTLVATDD94 pKa = 3.76AEE96 pKa = 5.01GNDD99 pKa = 4.14SVPTTAGFTGDD110 pKa = 4.14DD111 pKa = 3.91TPPTILIGALSGPTSGTYY129 pKa = 9.37IAAITLSEE137 pKa = 4.72DD138 pKa = 3.33STDD141 pKa = 4.5FDD143 pKa = 5.04AADD146 pKa = 3.69LTLTNATATLTGSGSDD162 pKa = 3.54YY163 pKa = 10.81TATLTPAADD172 pKa = 3.84GTVSLSVAAGVFTDD186 pKa = 4.59AAGNSNGASNTVSAVFAGVAPTVSISGAPEE216 pKa = 3.68ALAGGSSFTVAIAFSEE232 pKa = 4.68VVTGFVAADD241 pKa = 3.61ISATNATVTALTGGGVSYY259 pKa = 10.92VATVHH264 pKa = 6.95ASGTGDD270 pKa = 3.23VRR272 pKa = 11.84VAVPANVAFGATGHH286 pKa = 6.49GNLASSQVVIADD298 pKa = 3.22VSVQQTQEE306 pKa = 4.22HH307 pKa = 6.42IASYY311 pKa = 8.62MQTRR315 pKa = 11.84ANQLISNQPSLMSFLSGEE333 pKa = 4.06PRR335 pKa = 11.84GKK337 pKa = 10.77FNFAVTRR344 pKa = 11.84GAGSFDD350 pKa = 3.57VATGATGARR359 pKa = 11.84YY360 pKa = 8.2PVWAQANGSWTNDD373 pKa = 2.62GDD375 pKa = 4.23SEE377 pKa = 4.58SEE379 pKa = 4.46YY380 pKa = 11.33VFGALGGHH388 pKa = 4.73QTISEE393 pKa = 4.13NLLVGAMLQFDD404 pKa = 4.68HH405 pKa = 7.2LSEE408 pKa = 5.47DD409 pKa = 3.43EE410 pKa = 4.41GVASVRR416 pKa = 11.84GTGWMVGPYY425 pKa = 9.53FVARR429 pKa = 11.84SATQPLYY436 pKa = 10.91FEE438 pKa = 5.18GRR440 pKa = 11.84LLYY443 pKa = 11.05GEE445 pKa = 4.48TSNKK449 pKa = 9.32ISPFGTYY456 pKa = 9.63EE457 pKa = 4.1DD458 pKa = 4.53SFDD461 pKa = 3.74TTRR464 pKa = 11.84LLAQLKK470 pKa = 9.6VAGEE474 pKa = 4.08LSYY477 pKa = 11.44GRR479 pKa = 11.84TLLTPFMDD487 pKa = 4.11ASYY490 pKa = 9.26STDD493 pKa = 3.47DD494 pKa = 3.1QHH496 pKa = 9.94SYY498 pKa = 11.18VDD500 pKa = 3.75SLGNTIPGQDD510 pKa = 2.95IALGQIEE517 pKa = 4.42IGMDD521 pKa = 3.97FSMMLPVTTGDD532 pKa = 3.41LEE534 pKa = 4.27LWGGISGIWSHH545 pKa = 7.35IDD547 pKa = 3.08GSGYY551 pKa = 10.95ASTVTPDD558 pKa = 3.44YY559 pKa = 10.22EE560 pKa = 4.22GGRR563 pKa = 11.84ARR565 pKa = 11.84VEE567 pKa = 3.83LGINHH572 pKa = 6.26QDD574 pKa = 3.43SAGHH578 pKa = 6.2NFTVGTYY585 pKa = 10.24YY586 pKa = 10.9DD587 pKa = 4.23GIGANGYY594 pKa = 9.24EE595 pKa = 4.38SYY597 pKa = 11.06GLSLGYY603 pKa = 10.59EE604 pKa = 4.11MQFF607 pKa = 3.35
Molecular weight: 62.0 kDa
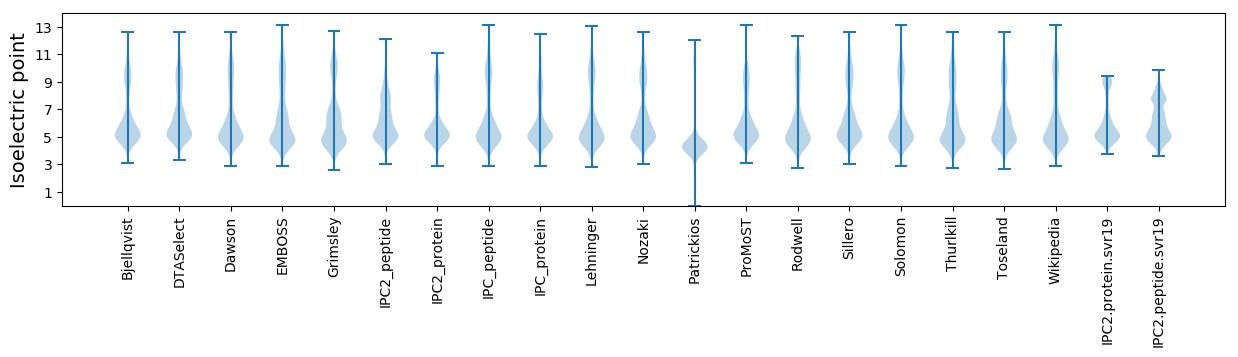
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2V4MNR1|A0A2V4MNR1_9RHOB Enoyl-CoA hydratase OS=Litorivita pollutaquae OX=2200892 GN=DI396_05095 PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 8.96RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.62GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.45AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 8.96RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.62GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.45AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
Molecular weight: 5.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
989163 |
28 |
1826 |
311.9 |
33.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.632 ± 0.06 | 0.889 ± 0.013 |
6.1 ± 0.042 | 5.883 ± 0.039 |
3.669 ± 0.026 | 8.728 ± 0.044 |
2.068 ± 0.02 | 5.335 ± 0.029 |
3.47 ± 0.038 | 9.677 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.953 ± 0.023 | 2.666 ± 0.022 |
4.876 ± 0.028 | 3.183 ± 0.023 |
6.466 ± 0.042 | 5.195 ± 0.027 |
5.527 ± 0.03 | 7.128 ± 0.034 |
1.274 ± 0.018 | 2.28 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
