
Spongiibacter sp. IMCC21906
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Cellvibrionales; Spongiibacteraceae; Spongiibacter; unclassified Spongiibacter
Average proteome isoelectric point is 6.15
Get precalculated fractions of proteins
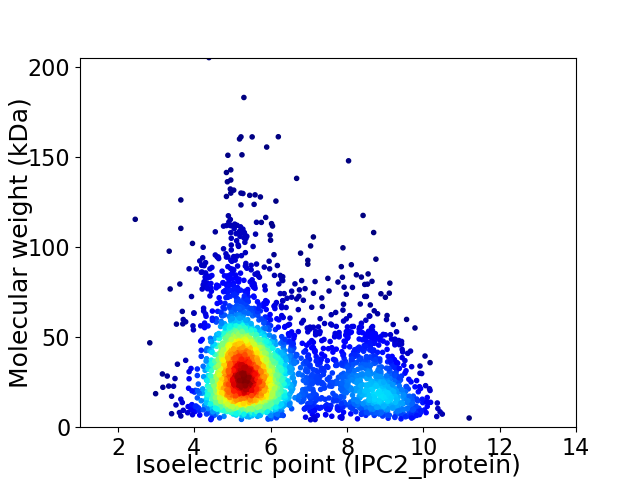
Virtual 2D-PAGE plot for 3192 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0F7M9B3|A0A0F7M9B3_9GAMM N-acetylmuramoyl-L-alanine amidase OS=Spongiibacter sp. IMCC21906 OX=1620392 GN=IMCC21906_03079 PE=4 SV=1
MM1 pKa = 7.61KK2 pKa = 10.53KK3 pKa = 10.33LFALSAISSALLLAACGGGGDD24 pKa = 4.54VNINAQNNSTGNGGGGGDD42 pKa = 3.43PGQTNSCASYY52 pKa = 11.19SDD54 pKa = 5.11DD55 pKa = 4.8NGTTRR60 pKa = 11.84QGSLDD65 pKa = 3.65GVNCVYY71 pKa = 9.97GTDD74 pKa = 4.3FVSLTNPIDD83 pKa = 3.49IGPSSRR89 pKa = 11.84VTFVKK94 pKa = 10.35LANNGVHH101 pKa = 5.97VFRR104 pKa = 11.84DD105 pKa = 3.53SLAVGKK111 pKa = 10.39NYY113 pKa = 10.87DD114 pKa = 3.68NDD116 pKa = 4.09ADD118 pKa = 3.75MAAAGISEE126 pKa = 4.66GGDD129 pKa = 3.18GSIIQIEE136 pKa = 4.26AGATLAFQSSDD147 pKa = 2.95DD148 pKa = 3.64YY149 pKa = 11.38MVINRR154 pKa = 11.84GSQIFAEE161 pKa = 4.92GAMNDD166 pKa = 4.45PITITSVSDD175 pKa = 3.35AVEE178 pKa = 3.98NSVDD182 pKa = 4.09PEE184 pKa = 5.02DD185 pKa = 3.73VQQWGGLIINGFGVTNKK202 pKa = 9.82CAYY205 pKa = 8.23TGSVDD210 pKa = 3.98GGDD213 pKa = 4.06LATSDD218 pKa = 3.7CHH220 pKa = 6.83VAAEE224 pKa = 4.59GKK226 pKa = 10.23AGAGQTHH233 pKa = 6.36YY234 pKa = 11.28GGDD237 pKa = 3.7NNEE240 pKa = 4.61DD241 pKa = 3.22NSGVLKK247 pKa = 10.48YY248 pKa = 9.67FIVKK252 pKa = 8.38HH253 pKa = 5.62TGAQVEE259 pKa = 4.61EE260 pKa = 4.35DD261 pKa = 3.71NEE263 pKa = 4.41LNGIAFDD270 pKa = 4.07AVGSEE275 pKa = 4.12TKK277 pKa = 10.3VDD279 pKa = 3.62YY280 pKa = 10.74LQAYY284 pKa = 6.86ATYY287 pKa = 10.64DD288 pKa = 3.26DD289 pKa = 5.04GIEE292 pKa = 4.1MFGGAVNISHH302 pKa = 6.44YY303 pKa = 10.24VALYY307 pKa = 10.98VNDD310 pKa = 4.29DD311 pKa = 4.37SIDD314 pKa = 3.37IDD316 pKa = 3.6EE317 pKa = 5.64GYY319 pKa = 10.62QGTIDD324 pKa = 3.84YY325 pKa = 10.9ALVIQSEE332 pKa = 4.65TDD334 pKa = 3.02GNRR337 pKa = 11.84CVEE340 pKa = 4.01SDD342 pKa = 3.7GVGSYY347 pKa = 10.83DD348 pKa = 3.9GRR350 pKa = 11.84TDD352 pKa = 3.33VADD355 pKa = 5.04LISRR359 pKa = 11.84GLNSRR364 pKa = 11.84ATIKK368 pKa = 10.85NLTCIISPSQVSGKK382 pKa = 10.65ADD384 pKa = 3.07INGADD389 pKa = 3.49GTGTHH394 pKa = 6.55EE395 pKa = 4.37EE396 pKa = 4.44GQGLRR401 pKa = 11.84IRR403 pKa = 11.84EE404 pKa = 3.97AHH406 pKa = 6.36YY407 pKa = 8.43PTISNALVTTAYY419 pKa = 10.44QADD422 pKa = 4.02QAPDD426 pKa = 3.26AANWCLRR433 pKa = 11.84IDD435 pKa = 4.01NEE437 pKa = 4.39GEE439 pKa = 3.82RR440 pKa = 11.84AALEE444 pKa = 4.13GDD446 pKa = 3.77LVIEE450 pKa = 4.02QSIFACQDD458 pKa = 3.03LTDD461 pKa = 5.0GDD463 pKa = 4.79DD464 pKa = 4.04LDD466 pKa = 4.5GTGTTQEE473 pKa = 4.27TFLTDD478 pKa = 2.97SGNQIYY484 pKa = 10.3QSLPEE489 pKa = 5.17GEE491 pKa = 5.15DD492 pKa = 3.23PTSASNADD500 pKa = 3.71LEE502 pKa = 4.45ILNGFYY508 pKa = 10.85SLLVGDD514 pKa = 4.38MVVGGAAATVTPTEE528 pKa = 3.61GRR530 pKa = 11.84AFVGAVTADD539 pKa = 4.1DD540 pKa = 4.12DD541 pKa = 4.17WTAGWAYY548 pKa = 10.57GLDD551 pKa = 3.62PANRR555 pKa = 11.84GQALWFEE562 pKa = 4.62
MM1 pKa = 7.61KK2 pKa = 10.53KK3 pKa = 10.33LFALSAISSALLLAACGGGGDD24 pKa = 4.54VNINAQNNSTGNGGGGGDD42 pKa = 3.43PGQTNSCASYY52 pKa = 11.19SDD54 pKa = 5.11DD55 pKa = 4.8NGTTRR60 pKa = 11.84QGSLDD65 pKa = 3.65GVNCVYY71 pKa = 9.97GTDD74 pKa = 4.3FVSLTNPIDD83 pKa = 3.49IGPSSRR89 pKa = 11.84VTFVKK94 pKa = 10.35LANNGVHH101 pKa = 5.97VFRR104 pKa = 11.84DD105 pKa = 3.53SLAVGKK111 pKa = 10.39NYY113 pKa = 10.87DD114 pKa = 3.68NDD116 pKa = 4.09ADD118 pKa = 3.75MAAAGISEE126 pKa = 4.66GGDD129 pKa = 3.18GSIIQIEE136 pKa = 4.26AGATLAFQSSDD147 pKa = 2.95DD148 pKa = 3.64YY149 pKa = 11.38MVINRR154 pKa = 11.84GSQIFAEE161 pKa = 4.92GAMNDD166 pKa = 4.45PITITSVSDD175 pKa = 3.35AVEE178 pKa = 3.98NSVDD182 pKa = 4.09PEE184 pKa = 5.02DD185 pKa = 3.73VQQWGGLIINGFGVTNKK202 pKa = 9.82CAYY205 pKa = 8.23TGSVDD210 pKa = 3.98GGDD213 pKa = 4.06LATSDD218 pKa = 3.7CHH220 pKa = 6.83VAAEE224 pKa = 4.59GKK226 pKa = 10.23AGAGQTHH233 pKa = 6.36YY234 pKa = 11.28GGDD237 pKa = 3.7NNEE240 pKa = 4.61DD241 pKa = 3.22NSGVLKK247 pKa = 10.48YY248 pKa = 9.67FIVKK252 pKa = 8.38HH253 pKa = 5.62TGAQVEE259 pKa = 4.61EE260 pKa = 4.35DD261 pKa = 3.71NEE263 pKa = 4.41LNGIAFDD270 pKa = 4.07AVGSEE275 pKa = 4.12TKK277 pKa = 10.3VDD279 pKa = 3.62YY280 pKa = 10.74LQAYY284 pKa = 6.86ATYY287 pKa = 10.64DD288 pKa = 3.26DD289 pKa = 5.04GIEE292 pKa = 4.1MFGGAVNISHH302 pKa = 6.44YY303 pKa = 10.24VALYY307 pKa = 10.98VNDD310 pKa = 4.29DD311 pKa = 4.37SIDD314 pKa = 3.37IDD316 pKa = 3.6EE317 pKa = 5.64GYY319 pKa = 10.62QGTIDD324 pKa = 3.84YY325 pKa = 10.9ALVIQSEE332 pKa = 4.65TDD334 pKa = 3.02GNRR337 pKa = 11.84CVEE340 pKa = 4.01SDD342 pKa = 3.7GVGSYY347 pKa = 10.83DD348 pKa = 3.9GRR350 pKa = 11.84TDD352 pKa = 3.33VADD355 pKa = 5.04LISRR359 pKa = 11.84GLNSRR364 pKa = 11.84ATIKK368 pKa = 10.85NLTCIISPSQVSGKK382 pKa = 10.65ADD384 pKa = 3.07INGADD389 pKa = 3.49GTGTHH394 pKa = 6.55EE395 pKa = 4.37EE396 pKa = 4.44GQGLRR401 pKa = 11.84IRR403 pKa = 11.84EE404 pKa = 3.97AHH406 pKa = 6.36YY407 pKa = 8.43PTISNALVTTAYY419 pKa = 10.44QADD422 pKa = 4.02QAPDD426 pKa = 3.26AANWCLRR433 pKa = 11.84IDD435 pKa = 4.01NEE437 pKa = 4.39GEE439 pKa = 3.82RR440 pKa = 11.84AALEE444 pKa = 4.13GDD446 pKa = 3.77LVIEE450 pKa = 4.02QSIFACQDD458 pKa = 3.03LTDD461 pKa = 5.0GDD463 pKa = 4.79DD464 pKa = 4.04LDD466 pKa = 4.5GTGTTQEE473 pKa = 4.27TFLTDD478 pKa = 2.97SGNQIYY484 pKa = 10.3QSLPEE489 pKa = 5.17GEE491 pKa = 5.15DD492 pKa = 3.23PTSASNADD500 pKa = 3.71LEE502 pKa = 4.45ILNGFYY508 pKa = 10.85SLLVGDD514 pKa = 4.38MVVGGAAATVTPTEE528 pKa = 3.61GRR530 pKa = 11.84AFVGAVTADD539 pKa = 4.1DD540 pKa = 4.12DD541 pKa = 4.17WTAGWAYY548 pKa = 10.57GLDD551 pKa = 3.62PANRR555 pKa = 11.84GQALWFEE562 pKa = 4.62
Molecular weight: 58.53 kDa
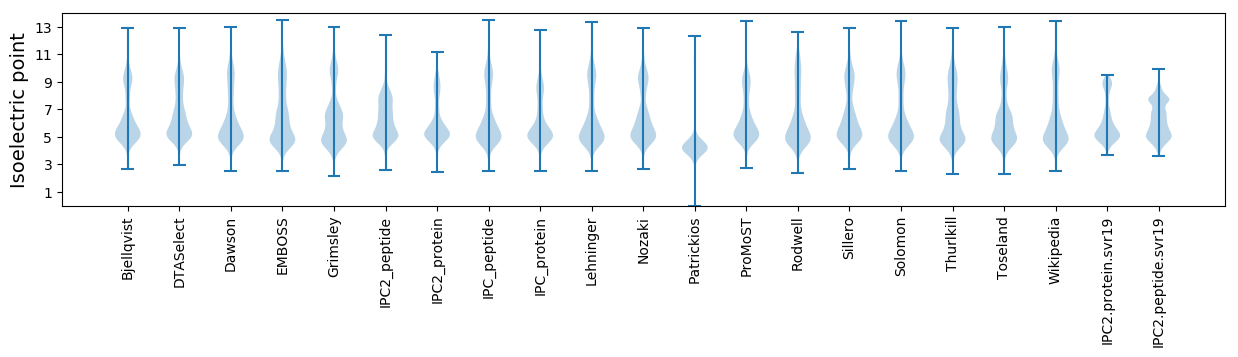
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0F7M1D5|A0A0F7M1D5_9GAMM Acetyltransferase ribosomal protein N-acetylase OS=Spongiibacter sp. IMCC21906 OX=1620392 GN=IMCC21906_00380 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.08RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.92GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.45NGRR28 pKa = 11.84KK29 pKa = 9.38LINRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.47GRR39 pKa = 11.84ASLSAA44 pKa = 3.83
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.08RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.92GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.45NGRR28 pKa = 11.84KK29 pKa = 9.38LINRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.47GRR39 pKa = 11.84ASLSAA44 pKa = 3.83
Molecular weight: 5.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1018612 |
39 |
1878 |
319.1 |
35.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.913 ± 0.052 | 1.086 ± 0.016 |
5.82 ± 0.037 | 6.12 ± 0.043 |
3.81 ± 0.029 | 7.502 ± 0.041 |
2.168 ± 0.022 | 5.615 ± 0.032 |
4.31 ± 0.035 | 10.807 ± 0.063 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.483 ± 0.023 | 3.666 ± 0.026 |
4.384 ± 0.031 | 4.386 ± 0.037 |
5.678 ± 0.033 | 6.602 ± 0.033 |
4.736 ± 0.031 | 6.841 ± 0.041 |
1.289 ± 0.017 | 2.785 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
