
Streptomyces taklimakanensis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces
Average proteome isoelectric point is 6.37
Get precalculated fractions of proteins
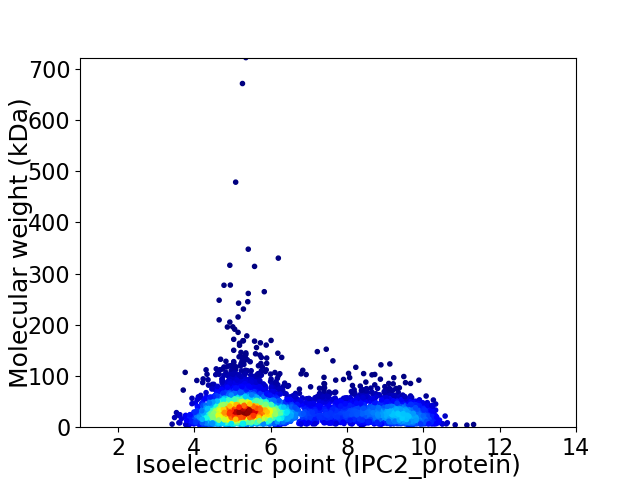
Virtual 2D-PAGE plot for 5063 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6G2B5V7|A0A6G2B5V7_9ACTN PIG-L family deacetylase OS=Streptomyces taklimakanensis OX=2569853 GN=F0L17_00520 PE=4 SV=1
MM1 pKa = 7.09KK2 pKa = 10.3RR3 pKa = 11.84RR4 pKa = 11.84WARR7 pKa = 11.84GLVAGAGVLALTGTSTLLMAAPAGAAIGNDD37 pKa = 3.24SNANAMASALAGPGADD53 pKa = 3.08VLGAGFEE60 pKa = 4.41SLAGAVANGVGDD72 pKa = 3.97SPLSNFPTNGSTFTILTSGDD92 pKa = 3.34AASADD97 pKa = 3.8DD98 pKa = 4.76PNTEE102 pKa = 4.15GSKK105 pKa = 7.7TTDD108 pKa = 3.26LAGGNVRR115 pKa = 11.84GNTDD119 pKa = 3.43FDD121 pKa = 4.26VSVFTVDD128 pKa = 3.51LQVPEE133 pKa = 4.3TANCLSFDD141 pKa = 3.57FQFYY145 pKa = 10.37SEE147 pKa = 5.59EE148 pKa = 4.0YY149 pKa = 9.03PEE151 pKa = 4.38WVGSAFNDD159 pKa = 3.43AFIAEE164 pKa = 5.07LDD166 pKa = 3.73NSDD169 pKa = 3.09WTTSGSEE176 pKa = 3.56INAPNNFAFDD186 pKa = 3.74ADD188 pKa = 3.77NDD190 pKa = 4.23VVSVNAVGLGGFSAEE205 pKa = 4.07NAAGTTYY212 pKa = 11.12DD213 pKa = 3.39GATPLLSAAKK223 pKa = 9.38QVTPGTHH230 pKa = 5.21TLYY233 pKa = 11.11LSIFDD238 pKa = 3.53QGDD241 pKa = 3.8RR242 pKa = 11.84ILDD245 pKa = 3.48SATFVDD251 pKa = 3.88NLRR254 pKa = 11.84VGFVPDD260 pKa = 5.02PEE262 pKa = 4.58TQCKK266 pKa = 10.24SGAQPKK272 pKa = 9.78PYY274 pKa = 10.07QLDD277 pKa = 3.7LTPVEE282 pKa = 4.5TTLDD286 pKa = 3.33TGTDD290 pKa = 3.42HH291 pKa = 6.99TVTATLTEE299 pKa = 4.38IGAEE303 pKa = 4.07PPAVSGASILFQASGAHH320 pKa = 5.98AVQGSDD326 pKa = 3.08TTDD329 pKa = 2.99TAGQATFTYY338 pKa = 9.39TGTTVGVDD346 pKa = 4.0SISACYY352 pKa = 10.19DD353 pKa = 2.74VDD355 pKa = 4.85GNGTCDD361 pKa = 3.14AGEE364 pKa = 4.31PFASAKK370 pKa = 9.29ATWKK374 pKa = 8.07NTPPVNDD381 pKa = 4.0PGGPYY386 pKa = 10.2SGAEE390 pKa = 4.14GSAVALDD397 pKa = 3.82ATASDD402 pKa = 4.22VNGDD406 pKa = 4.0PLTHH410 pKa = 6.55KK411 pKa = 8.22WTYY414 pKa = 10.33EE415 pKa = 3.85PVSGVDD421 pKa = 3.28TGAVCSFEE429 pKa = 5.45DD430 pKa = 4.86DD431 pKa = 3.88SALEE435 pKa = 3.95TAITCTDD442 pKa = 3.47DD443 pKa = 3.54GEE445 pKa = 4.43YY446 pKa = 10.82RR447 pKa = 11.84LTLTTDD453 pKa = 3.54DD454 pKa = 4.25GVNAPVSNTADD465 pKa = 3.43LTVTNVAPAIGEE477 pKa = 3.97VSTPVDD483 pKa = 3.67PVAVGTPLSLEE494 pKa = 4.1AGYY497 pKa = 8.99TDD499 pKa = 4.3PGANDD504 pKa = 3.26THH506 pKa = 6.33TASIDD511 pKa = 3.3WGDD514 pKa = 3.53GSMSEE519 pKa = 4.08PTAAAGQVDD528 pKa = 4.0ASHH531 pKa = 7.32TYY533 pKa = 8.19TAAGIYY539 pKa = 7.27TICLTVTDD547 pKa = 5.21DD548 pKa = 6.07DD549 pKa = 4.43GASDD553 pKa = 4.18EE554 pKa = 4.6KK555 pKa = 11.08CSPNYY560 pKa = 10.05VVVYY564 pKa = 9.98DD565 pKa = 4.69PSAGFVTGGGWIDD578 pKa = 3.75VPEE581 pKa = 4.53GSYY584 pKa = 10.43PADD587 pKa = 3.9PSASGPGRR595 pKa = 11.84FGFVSKK601 pKa = 9.63YY602 pKa = 8.8QKK604 pKa = 10.81GATAPKK610 pKa = 9.95GQTEE614 pKa = 4.45FQFKK618 pKa = 9.47TGSLNFHH625 pKa = 6.38SMSYY629 pKa = 10.8DD630 pKa = 2.94WLVVSGNKK638 pKa = 9.81AIYY641 pKa = 10.01KK642 pKa = 8.43GTGTVNGEE650 pKa = 3.92DD651 pKa = 3.86GYY653 pKa = 11.5GFLVSVVDD661 pKa = 3.97ADD663 pKa = 4.26KK664 pKa = 11.58SGGDD668 pKa = 3.07DD669 pKa = 3.46TYY671 pKa = 11.14RR672 pKa = 11.84IKK674 pKa = 10.96VWDD677 pKa = 4.27ADD679 pKa = 3.38SDD681 pKa = 4.33TVVFDD686 pKa = 4.2NQPGAADD693 pKa = 3.61DD694 pKa = 4.08TAATTAISHH703 pKa = 6.19GSIVIHH709 pKa = 6.2SS710 pKa = 3.78
MM1 pKa = 7.09KK2 pKa = 10.3RR3 pKa = 11.84RR4 pKa = 11.84WARR7 pKa = 11.84GLVAGAGVLALTGTSTLLMAAPAGAAIGNDD37 pKa = 3.24SNANAMASALAGPGADD53 pKa = 3.08VLGAGFEE60 pKa = 4.41SLAGAVANGVGDD72 pKa = 3.97SPLSNFPTNGSTFTILTSGDD92 pKa = 3.34AASADD97 pKa = 3.8DD98 pKa = 4.76PNTEE102 pKa = 4.15GSKK105 pKa = 7.7TTDD108 pKa = 3.26LAGGNVRR115 pKa = 11.84GNTDD119 pKa = 3.43FDD121 pKa = 4.26VSVFTVDD128 pKa = 3.51LQVPEE133 pKa = 4.3TANCLSFDD141 pKa = 3.57FQFYY145 pKa = 10.37SEE147 pKa = 5.59EE148 pKa = 4.0YY149 pKa = 9.03PEE151 pKa = 4.38WVGSAFNDD159 pKa = 3.43AFIAEE164 pKa = 5.07LDD166 pKa = 3.73NSDD169 pKa = 3.09WTTSGSEE176 pKa = 3.56INAPNNFAFDD186 pKa = 3.74ADD188 pKa = 3.77NDD190 pKa = 4.23VVSVNAVGLGGFSAEE205 pKa = 4.07NAAGTTYY212 pKa = 11.12DD213 pKa = 3.39GATPLLSAAKK223 pKa = 9.38QVTPGTHH230 pKa = 5.21TLYY233 pKa = 11.11LSIFDD238 pKa = 3.53QGDD241 pKa = 3.8RR242 pKa = 11.84ILDD245 pKa = 3.48SATFVDD251 pKa = 3.88NLRR254 pKa = 11.84VGFVPDD260 pKa = 5.02PEE262 pKa = 4.58TQCKK266 pKa = 10.24SGAQPKK272 pKa = 9.78PYY274 pKa = 10.07QLDD277 pKa = 3.7LTPVEE282 pKa = 4.5TTLDD286 pKa = 3.33TGTDD290 pKa = 3.42HH291 pKa = 6.99TVTATLTEE299 pKa = 4.38IGAEE303 pKa = 4.07PPAVSGASILFQASGAHH320 pKa = 5.98AVQGSDD326 pKa = 3.08TTDD329 pKa = 2.99TAGQATFTYY338 pKa = 9.39TGTTVGVDD346 pKa = 4.0SISACYY352 pKa = 10.19DD353 pKa = 2.74VDD355 pKa = 4.85GNGTCDD361 pKa = 3.14AGEE364 pKa = 4.31PFASAKK370 pKa = 9.29ATWKK374 pKa = 8.07NTPPVNDD381 pKa = 4.0PGGPYY386 pKa = 10.2SGAEE390 pKa = 4.14GSAVALDD397 pKa = 3.82ATASDD402 pKa = 4.22VNGDD406 pKa = 4.0PLTHH410 pKa = 6.55KK411 pKa = 8.22WTYY414 pKa = 10.33EE415 pKa = 3.85PVSGVDD421 pKa = 3.28TGAVCSFEE429 pKa = 5.45DD430 pKa = 4.86DD431 pKa = 3.88SALEE435 pKa = 3.95TAITCTDD442 pKa = 3.47DD443 pKa = 3.54GEE445 pKa = 4.43YY446 pKa = 10.82RR447 pKa = 11.84LTLTTDD453 pKa = 3.54DD454 pKa = 4.25GVNAPVSNTADD465 pKa = 3.43LTVTNVAPAIGEE477 pKa = 3.97VSTPVDD483 pKa = 3.67PVAVGTPLSLEE494 pKa = 4.1AGYY497 pKa = 8.99TDD499 pKa = 4.3PGANDD504 pKa = 3.26THH506 pKa = 6.33TASIDD511 pKa = 3.3WGDD514 pKa = 3.53GSMSEE519 pKa = 4.08PTAAAGQVDD528 pKa = 4.0ASHH531 pKa = 7.32TYY533 pKa = 8.19TAAGIYY539 pKa = 7.27TICLTVTDD547 pKa = 5.21DD548 pKa = 6.07DD549 pKa = 4.43GASDD553 pKa = 4.18EE554 pKa = 4.6KK555 pKa = 11.08CSPNYY560 pKa = 10.05VVVYY564 pKa = 9.98DD565 pKa = 4.69PSAGFVTGGGWIDD578 pKa = 3.75VPEE581 pKa = 4.53GSYY584 pKa = 10.43PADD587 pKa = 3.9PSASGPGRR595 pKa = 11.84FGFVSKK601 pKa = 9.63YY602 pKa = 8.8QKK604 pKa = 10.81GATAPKK610 pKa = 9.95GQTEE614 pKa = 4.45FQFKK618 pKa = 9.47TGSLNFHH625 pKa = 6.38SMSYY629 pKa = 10.8DD630 pKa = 2.94WLVVSGNKK638 pKa = 9.81AIYY641 pKa = 10.01KK642 pKa = 8.43GTGTVNGEE650 pKa = 3.92DD651 pKa = 3.86GYY653 pKa = 11.5GFLVSVVDD661 pKa = 3.97ADD663 pKa = 4.26KK664 pKa = 11.58SGGDD668 pKa = 3.07DD669 pKa = 3.46TYY671 pKa = 11.14RR672 pKa = 11.84IKK674 pKa = 10.96VWDD677 pKa = 4.27ADD679 pKa = 3.38SDD681 pKa = 4.33TVVFDD686 pKa = 4.2NQPGAADD693 pKa = 3.61DD694 pKa = 4.08TAATTAISHH703 pKa = 6.19GSIVIHH709 pKa = 6.2SS710 pKa = 3.78
Molecular weight: 72.51 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6G2BDV9|A0A6G2BDV9_9ACTN Uncharacterized protein OS=Streptomyces taklimakanensis OX=2569853 GN=F0L17_14605 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AIIASRR35 pKa = 11.84RR36 pKa = 11.84GKK38 pKa = 10.34GRR40 pKa = 11.84ARR42 pKa = 11.84LSAA45 pKa = 3.91
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AIIASRR35 pKa = 11.84RR36 pKa = 11.84GKK38 pKa = 10.34GRR40 pKa = 11.84ARR42 pKa = 11.84LSAA45 pKa = 3.91
Molecular weight: 5.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1706788 |
32 |
6685 |
337.1 |
36.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.314 ± 0.057 | 0.8 ± 0.01 |
6.121 ± 0.029 | 6.347 ± 0.032 |
2.581 ± 0.016 | 9.911 ± 0.035 |
2.356 ± 0.016 | 2.775 ± 0.026 |
1.764 ± 0.03 | 10.049 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.63 ± 0.014 | 1.639 ± 0.019 |
6.43 ± 0.036 | 2.232 ± 0.024 |
8.982 ± 0.042 | 4.761 ± 0.027 |
6.067 ± 0.026 | 8.791 ± 0.029 |
1.484 ± 0.015 | 1.966 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
