
Ateles paniscus polyomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; Alphapolyomavirus
Average proteome isoelectric point is 5.92
Get precalculated fractions of proteins
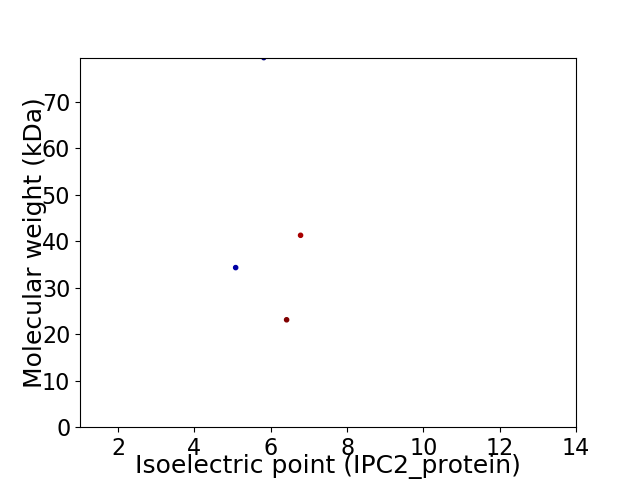
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K7QK07|K7QK07_9POLY Small T antigen OS=Ateles paniscus polyomavirus 1 OX=1236391 PE=4 SV=1
MM1 pKa = 7.46GGVLSLFFDD10 pKa = 3.5IAEE13 pKa = 4.41LASALSTEE21 pKa = 4.34TGISVGAILAGEE33 pKa = 4.09AAADD37 pKa = 3.55IEE39 pKa = 4.31ASVMGLMTIEE49 pKa = 4.18GLGAADD55 pKa = 4.18ALAAMGLTMEE65 pKa = 5.06NFSMMYY71 pKa = 9.88ALPGMLSEE79 pKa = 4.54AVGIGTLFQTISGASSLVAVGIKK102 pKa = 9.75YY103 pKa = 10.44SRR105 pKa = 11.84EE106 pKa = 3.62EE107 pKa = 3.66VSIVNKK113 pKa = 10.31NISDD117 pKa = 3.67MALQLWRR124 pKa = 11.84PQDD127 pKa = 3.56YY128 pKa = 11.22YY129 pKa = 11.46DD130 pKa = 3.73ILFPGVNHH138 pKa = 6.51FAHH141 pKa = 6.32YY142 pKa = 10.85LNVIEE147 pKa = 5.31GWASSLVHH155 pKa = 5.38TVSRR159 pKa = 11.84AIWDD163 pKa = 3.77SIMRR167 pKa = 11.84DD168 pKa = 3.06ARR170 pKa = 11.84HH171 pKa = 5.53QIGYY175 pKa = 10.39ASRR178 pKa = 11.84EE179 pKa = 3.96LVVRR183 pKa = 11.84GTTSFQDD190 pKa = 3.21VMAKK194 pKa = 10.3VIEE197 pKa = 4.04TSRR200 pKa = 11.84WVLTSGPSHH209 pKa = 7.14IYY211 pKa = 10.29SSLSEE216 pKa = 4.43YY217 pKa = 10.35YY218 pKa = 10.14RR219 pKa = 11.84QLPGLNPAQIRR230 pKa = 11.84DD231 pKa = 3.44VYY233 pKa = 10.69RR234 pKa = 11.84RR235 pKa = 11.84LGEE238 pKa = 4.11PVPSRR243 pKa = 11.84YY244 pKa = 8.92QLEE247 pKa = 4.18VEE249 pKa = 4.17ARR251 pKa = 11.84EE252 pKa = 4.02EE253 pKa = 3.98SAEE256 pKa = 4.14VIEE259 pKa = 5.07TYY261 pKa = 10.25SAPGGAHH268 pKa = 6.07QRR270 pKa = 11.84VCPDD274 pKa = 2.37WMLPLILGLYY284 pKa = 10.41GDD286 pKa = 4.05VTPTFATYY294 pKa = 9.37IHH296 pKa = 5.77QVEE299 pKa = 4.4KK300 pKa = 10.72EE301 pKa = 3.99DD302 pKa = 4.53DD303 pKa = 3.21KK304 pKa = 11.77GRR306 pKa = 11.84PSKK309 pKa = 10.45RR310 pKa = 11.84RR311 pKa = 11.84RR312 pKa = 11.84LL313 pKa = 3.55
MM1 pKa = 7.46GGVLSLFFDD10 pKa = 3.5IAEE13 pKa = 4.41LASALSTEE21 pKa = 4.34TGISVGAILAGEE33 pKa = 4.09AAADD37 pKa = 3.55IEE39 pKa = 4.31ASVMGLMTIEE49 pKa = 4.18GLGAADD55 pKa = 4.18ALAAMGLTMEE65 pKa = 5.06NFSMMYY71 pKa = 9.88ALPGMLSEE79 pKa = 4.54AVGIGTLFQTISGASSLVAVGIKK102 pKa = 9.75YY103 pKa = 10.44SRR105 pKa = 11.84EE106 pKa = 3.62EE107 pKa = 3.66VSIVNKK113 pKa = 10.31NISDD117 pKa = 3.67MALQLWRR124 pKa = 11.84PQDD127 pKa = 3.56YY128 pKa = 11.22YY129 pKa = 11.46DD130 pKa = 3.73ILFPGVNHH138 pKa = 6.51FAHH141 pKa = 6.32YY142 pKa = 10.85LNVIEE147 pKa = 5.31GWASSLVHH155 pKa = 5.38TVSRR159 pKa = 11.84AIWDD163 pKa = 3.77SIMRR167 pKa = 11.84DD168 pKa = 3.06ARR170 pKa = 11.84HH171 pKa = 5.53QIGYY175 pKa = 10.39ASRR178 pKa = 11.84EE179 pKa = 3.96LVVRR183 pKa = 11.84GTTSFQDD190 pKa = 3.21VMAKK194 pKa = 10.3VIEE197 pKa = 4.04TSRR200 pKa = 11.84WVLTSGPSHH209 pKa = 7.14IYY211 pKa = 10.29SSLSEE216 pKa = 4.43YY217 pKa = 10.35YY218 pKa = 10.14RR219 pKa = 11.84QLPGLNPAQIRR230 pKa = 11.84DD231 pKa = 3.44VYY233 pKa = 10.69RR234 pKa = 11.84RR235 pKa = 11.84LGEE238 pKa = 4.11PVPSRR243 pKa = 11.84YY244 pKa = 8.92QLEE247 pKa = 4.18VEE249 pKa = 4.17ARR251 pKa = 11.84EE252 pKa = 4.02EE253 pKa = 3.98SAEE256 pKa = 4.14VIEE259 pKa = 5.07TYY261 pKa = 10.25SAPGGAHH268 pKa = 6.07QRR270 pKa = 11.84VCPDD274 pKa = 2.37WMLPLILGLYY284 pKa = 10.41GDD286 pKa = 4.05VTPTFATYY294 pKa = 9.37IHH296 pKa = 5.77QVEE299 pKa = 4.4KK300 pKa = 10.72EE301 pKa = 3.99DD302 pKa = 4.53DD303 pKa = 3.21KK304 pKa = 11.77GRR306 pKa = 11.84PSKK309 pKa = 10.45RR310 pKa = 11.84RR311 pKa = 11.84RR312 pKa = 11.84LL313 pKa = 3.55
Molecular weight: 34.34 kDa
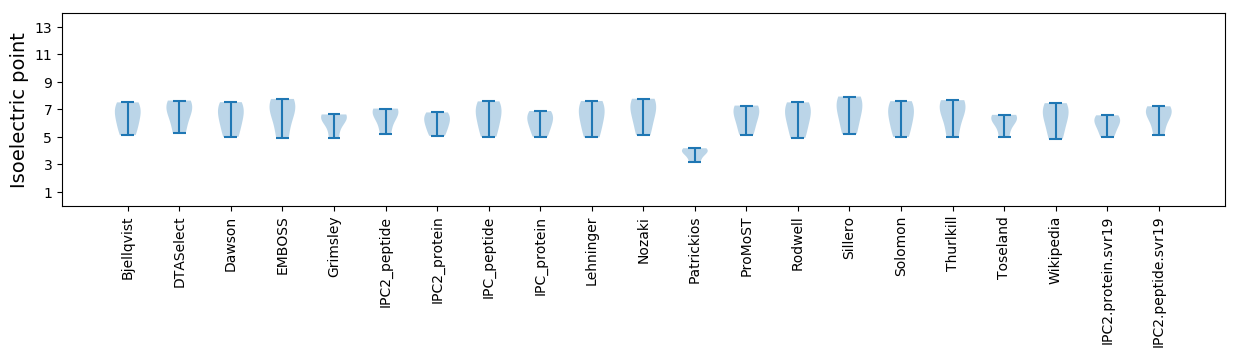
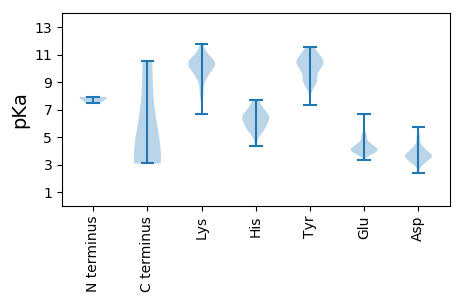
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K7QLH4|K7QLH4_9POLY Capsid protein VP1 OS=Ateles paniscus polyomavirus 1 OX=1236391 PE=3 SV=1
MM1 pKa = 7.71ASKK4 pKa = 10.36KK5 pKa = 10.09CPRR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 8.22TPSCPRR16 pKa = 11.84KK17 pKa = 9.06PATCPQTTCPVPRR30 pKa = 11.84AVPRR34 pKa = 11.84LLIKK38 pKa = 10.88GGVEE42 pKa = 3.73VLAVKK47 pKa = 9.34TGPDD51 pKa = 3.81SITQIEE57 pKa = 4.95AYY59 pKa = 10.36LNPRR63 pKa = 11.84MGKK66 pKa = 10.07AGNDD70 pKa = 3.43DD71 pKa = 3.54QYY73 pKa = 11.53GYY75 pKa = 10.19SASITVAASLDD86 pKa = 3.31ADD88 pKa = 3.73NPKK91 pKa = 10.27KK92 pKa = 10.73EE93 pKa = 4.17EE94 pKa = 4.07LPTYY98 pKa = 8.96STAKK102 pKa = 10.17ISLPLLNEE110 pKa = 4.58DD111 pKa = 4.2LTCGTLLMWEE121 pKa = 4.14AVSVKK126 pKa = 9.88TEE128 pKa = 3.84VVGISSLVNVHH139 pKa = 6.17IPSKK143 pKa = 10.69RR144 pKa = 11.84MYY146 pKa = 8.85NDD148 pKa = 2.99KK149 pKa = 11.3GIGFPIEE156 pKa = 3.8GMNFHH161 pKa = 6.75MFAVGGEE168 pKa = 4.05PLEE171 pKa = 4.35LQAVTSNYY179 pKa = 7.35KK180 pKa = 9.37TNYY183 pKa = 8.66PDD185 pKa = 3.58GTVKK189 pKa = 10.68PPVKK193 pKa = 10.57AKK195 pKa = 8.27TQQVLDD201 pKa = 3.95PQYY204 pKa = 10.88KK205 pKa = 10.44AKK207 pKa = 10.46LLKK210 pKa = 10.86DD211 pKa = 3.17GAYY214 pKa = 9.64PIEE217 pKa = 4.66LWAPDD222 pKa = 3.61PSMNEE227 pKa = 3.34NTRR230 pKa = 11.84YY231 pKa = 9.36YY232 pKa = 11.08GSYY235 pKa = 9.74TGGQSTPPVLQFTNTVTTVLLDD257 pKa = 3.84EE258 pKa = 5.12NGVGPLCKK266 pKa = 10.13GDD268 pKa = 4.16ALYY271 pKa = 8.95LTAADD276 pKa = 2.74IVGFHH281 pKa = 6.32TNAAEE286 pKa = 3.98TMAYY290 pKa = 9.9RR291 pKa = 11.84GLPRR295 pKa = 11.84YY296 pKa = 10.01FNVTLRR302 pKa = 11.84KK303 pKa = 9.36RR304 pKa = 11.84AVKK307 pKa = 10.4NPYY310 pKa = 9.15PVSTLLNNLFTGYY323 pKa = 9.98LPKK326 pKa = 10.56VEE328 pKa = 4.18GQPMNDD334 pKa = 3.13KK335 pKa = 10.66NGEE338 pKa = 4.03VGQVEE343 pKa = 4.47EE344 pKa = 4.15VRR346 pKa = 11.84VYY348 pKa = 11.12EE349 pKa = 4.42GMEE352 pKa = 4.1PTPGDD357 pKa = 3.56PDD359 pKa = 3.76MIRR362 pKa = 11.84YY363 pKa = 9.33LDD365 pKa = 3.61EE366 pKa = 5.19FGQQKK371 pKa = 9.02TSPNMNN377 pKa = 3.48
MM1 pKa = 7.71ASKK4 pKa = 10.36KK5 pKa = 10.09CPRR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 8.22TPSCPRR16 pKa = 11.84KK17 pKa = 9.06PATCPQTTCPVPRR30 pKa = 11.84AVPRR34 pKa = 11.84LLIKK38 pKa = 10.88GGVEE42 pKa = 3.73VLAVKK47 pKa = 9.34TGPDD51 pKa = 3.81SITQIEE57 pKa = 4.95AYY59 pKa = 10.36LNPRR63 pKa = 11.84MGKK66 pKa = 10.07AGNDD70 pKa = 3.43DD71 pKa = 3.54QYY73 pKa = 11.53GYY75 pKa = 10.19SASITVAASLDD86 pKa = 3.31ADD88 pKa = 3.73NPKK91 pKa = 10.27KK92 pKa = 10.73EE93 pKa = 4.17EE94 pKa = 4.07LPTYY98 pKa = 8.96STAKK102 pKa = 10.17ISLPLLNEE110 pKa = 4.58DD111 pKa = 4.2LTCGTLLMWEE121 pKa = 4.14AVSVKK126 pKa = 9.88TEE128 pKa = 3.84VVGISSLVNVHH139 pKa = 6.17IPSKK143 pKa = 10.69RR144 pKa = 11.84MYY146 pKa = 8.85NDD148 pKa = 2.99KK149 pKa = 11.3GIGFPIEE156 pKa = 3.8GMNFHH161 pKa = 6.75MFAVGGEE168 pKa = 4.05PLEE171 pKa = 4.35LQAVTSNYY179 pKa = 7.35KK180 pKa = 9.37TNYY183 pKa = 8.66PDD185 pKa = 3.58GTVKK189 pKa = 10.68PPVKK193 pKa = 10.57AKK195 pKa = 8.27TQQVLDD201 pKa = 3.95PQYY204 pKa = 10.88KK205 pKa = 10.44AKK207 pKa = 10.46LLKK210 pKa = 10.86DD211 pKa = 3.17GAYY214 pKa = 9.64PIEE217 pKa = 4.66LWAPDD222 pKa = 3.61PSMNEE227 pKa = 3.34NTRR230 pKa = 11.84YY231 pKa = 9.36YY232 pKa = 11.08GSYY235 pKa = 9.74TGGQSTPPVLQFTNTVTTVLLDD257 pKa = 3.84EE258 pKa = 5.12NGVGPLCKK266 pKa = 10.13GDD268 pKa = 4.16ALYY271 pKa = 8.95LTAADD276 pKa = 2.74IVGFHH281 pKa = 6.32TNAAEE286 pKa = 3.98TMAYY290 pKa = 9.9RR291 pKa = 11.84GLPRR295 pKa = 11.84YY296 pKa = 10.01FNVTLRR302 pKa = 11.84KK303 pKa = 9.36RR304 pKa = 11.84AVKK307 pKa = 10.4NPYY310 pKa = 9.15PVSTLLNNLFTGYY323 pKa = 9.98LPKK326 pKa = 10.56VEE328 pKa = 4.18GQPMNDD334 pKa = 3.13KK335 pKa = 10.66NGEE338 pKa = 4.03VGQVEE343 pKa = 4.47EE344 pKa = 4.15VRR346 pKa = 11.84VYY348 pKa = 11.12EE349 pKa = 4.42GMEE352 pKa = 4.1PTPGDD357 pKa = 3.56PDD359 pKa = 3.76MIRR362 pKa = 11.84YY363 pKa = 9.33LDD365 pKa = 3.61EE366 pKa = 5.19FGQQKK371 pKa = 9.02TSPNMNN377 pKa = 3.48
Molecular weight: 41.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1584 |
197 |
697 |
396.0 |
44.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.503 ± 0.814 | 2.525 ± 0.648 |
5.366 ± 0.394 | 6.755 ± 0.368 |
4.293 ± 0.794 | 6.692 ± 0.677 |
2.146 ± 0.392 | 4.419 ± 0.599 |
6.692 ± 1.15 | 9.848 ± 0.482 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.093 ± 0.366 | 4.545 ± 0.837 |
5.808 ± 1.006 | 3.535 ± 0.094 |
4.293 ± 0.417 | 6.566 ± 0.876 |
5.24 ± 0.839 | 6.187 ± 0.635 |
1.515 ± 0.397 | 3.977 ± 0.561 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
