
Eptesicus serotinus papillomavirus 2
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Dyoomegapapillomavirus; Dyoomegapapillomavirus 1
Average proteome isoelectric point is 5.83
Get precalculated fractions of proteins
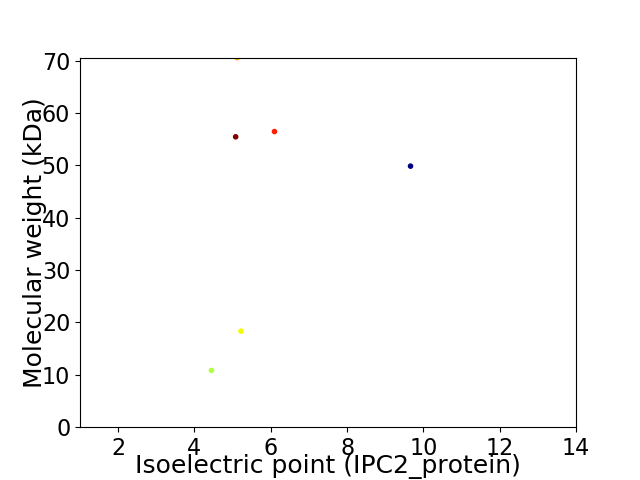
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W8EC56|W8EC56_9PAPI Protein E6 OS=Eptesicus serotinus papillomavirus 2 OX=1464072 GN=E6 PE=3 SV=1
MM1 pKa = 7.56IGNAPTLKK9 pKa = 10.64DD10 pKa = 3.18IVLTEE15 pKa = 3.8IPEE18 pKa = 4.3AVDD21 pKa = 3.94LLCHH25 pKa = 5.38EE26 pKa = 4.68TMPSEE31 pKa = 4.36EE32 pKa = 4.15EE33 pKa = 3.91LGAGDD38 pKa = 3.54SHH40 pKa = 8.32SGNRR44 pKa = 11.84PRR46 pKa = 11.84AQQVHH51 pKa = 6.8VILTWCGRR59 pKa = 11.84CDD61 pKa = 3.27QDD63 pKa = 3.89IRR65 pKa = 11.84VCIEE69 pKa = 3.79TEE71 pKa = 3.97QQSLVILEE79 pKa = 4.16SLLLKK84 pKa = 10.34DD85 pKa = 4.72LQILCPEE92 pKa = 5.23CYY94 pKa = 9.97QNLL97 pKa = 3.69
MM1 pKa = 7.56IGNAPTLKK9 pKa = 10.64DD10 pKa = 3.18IVLTEE15 pKa = 3.8IPEE18 pKa = 4.3AVDD21 pKa = 3.94LLCHH25 pKa = 5.38EE26 pKa = 4.68TMPSEE31 pKa = 4.36EE32 pKa = 4.15EE33 pKa = 3.91LGAGDD38 pKa = 3.54SHH40 pKa = 8.32SGNRR44 pKa = 11.84PRR46 pKa = 11.84AQQVHH51 pKa = 6.8VILTWCGRR59 pKa = 11.84CDD61 pKa = 3.27QDD63 pKa = 3.89IRR65 pKa = 11.84VCIEE69 pKa = 3.79TEE71 pKa = 3.97QQSLVILEE79 pKa = 4.16SLLLKK84 pKa = 10.34DD85 pKa = 4.72LQILCPEE92 pKa = 5.23CYY94 pKa = 9.97QNLL97 pKa = 3.69
Molecular weight: 10.84 kDa
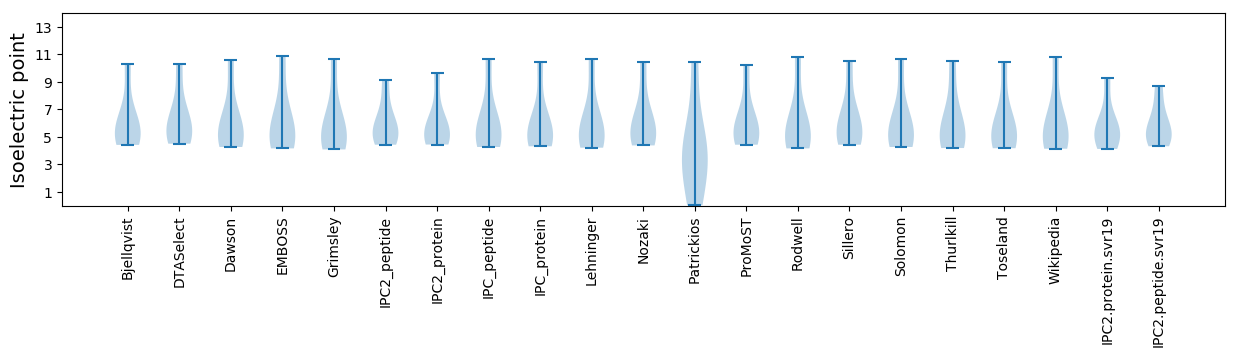
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W8EH55|W8EH55_9PAPI Regulatory protein E2 OS=Eptesicus serotinus papillomavirus 2 OX=1464072 GN=E2 PE=3 SV=1
MM1 pKa = 7.46KK2 pKa = 10.25RR3 pKa = 11.84RR4 pKa = 11.84EE5 pKa = 4.1TTEE8 pKa = 3.74QLTSRR13 pKa = 11.84LDD15 pKa = 3.48AVQEE19 pKa = 4.1KK20 pKa = 10.18ILAHH24 pKa = 6.48FEE26 pKa = 4.26TTSTEE31 pKa = 3.84LAAHH35 pKa = 6.34IEE37 pKa = 4.17YY38 pKa = 9.68WSLVRR43 pKa = 11.84QEE45 pKa = 4.91SVILYY50 pKa = 6.66YY51 pKa = 10.67ARR53 pKa = 11.84HH54 pKa = 6.05LGVNRR59 pKa = 11.84LGLQPVPAQAISEE72 pKa = 4.15GRR74 pKa = 11.84AKK76 pKa = 10.16QAIGMQLVLEE86 pKa = 4.27SLQRR90 pKa = 11.84SPYY93 pKa = 10.36ARR95 pKa = 11.84EE96 pKa = 3.39EE97 pKa = 3.82WTLQDD102 pKa = 3.49VSLEE106 pKa = 4.22KK107 pKa = 11.15YY108 pKa = 10.13SVQPKK113 pKa = 8.58NTFKK117 pKa = 11.03KK118 pKa = 10.45DD119 pKa = 3.08GTQVHH124 pKa = 5.8VKK126 pKa = 10.18YY127 pKa = 10.86GPDD130 pKa = 3.3PTDD133 pKa = 3.77FMPYY137 pKa = 9.77VLWGSIYY144 pKa = 10.69YY145 pKa = 10.02QDD147 pKa = 5.82VDD149 pKa = 4.52DD150 pKa = 4.97KK151 pKa = 9.07WHH153 pKa = 6.45KK154 pKa = 11.03VPGQVDD160 pKa = 3.53YY161 pKa = 11.49HH162 pKa = 7.07GLFYY166 pKa = 10.61MEE168 pKa = 4.6GKK170 pKa = 10.12EE171 pKa = 4.03KK172 pKa = 9.89IYY174 pKa = 10.96YY175 pKa = 10.39EE176 pKa = 4.09NFKK179 pKa = 10.88EE180 pKa = 4.0DD181 pKa = 3.86AIKK184 pKa = 10.62FGMPGRR190 pKa = 11.84WIVNVKK196 pKa = 8.4NTAVPSFSRR205 pKa = 11.84QTSSAFAGGRR215 pKa = 11.84PGADD219 pKa = 2.94STDD222 pKa = 3.47APEE225 pKa = 4.61APSTSRR231 pKa = 11.84WSPRR235 pKa = 11.84ASNTTQAQEE244 pKa = 4.34SNQVSGPSTRR254 pKa = 11.84RR255 pKa = 11.84GRR257 pKa = 11.84GRR259 pKa = 11.84RR260 pKa = 11.84SPSPGEE266 pKa = 4.26GPSSSRR272 pKa = 11.84RR273 pKa = 11.84RR274 pKa = 11.84RR275 pKa = 11.84LPTAARR281 pKa = 11.84RR282 pKa = 11.84TPSPEE287 pKa = 3.2RR288 pKa = 11.84HH289 pKa = 5.36SRR291 pKa = 11.84SRR293 pKa = 11.84SRR295 pKa = 11.84DD296 pKa = 2.81WRR298 pKa = 11.84GGRR301 pKa = 11.84GGGGGLRR308 pKa = 11.84GRR310 pKa = 11.84GRR312 pKa = 11.84QGEE315 pKa = 4.44SAGRR319 pKa = 11.84SAGPHH324 pKa = 5.39SRR326 pKa = 11.84SSQSPAPTPPTPGQVGGRR344 pKa = 11.84HH345 pKa = 4.63RR346 pKa = 11.84TAEE349 pKa = 4.22PGASRR354 pKa = 11.84LEE356 pKa = 4.01RR357 pKa = 11.84LIQEE361 pKa = 4.31ARR363 pKa = 11.84DD364 pKa = 3.7PPVVLIRR371 pKa = 11.84GTPNGVKK378 pKa = 10.2CLRR381 pKa = 11.84YY382 pKa = 9.56RR383 pKa = 11.84LRR385 pKa = 11.84TQNQHH390 pKa = 6.65LYY392 pKa = 10.99DD393 pKa = 4.01SVTSTFFWVNKK404 pKa = 9.72NGKK407 pKa = 6.35QQEE410 pKa = 4.23GRR412 pKa = 11.84IMLSFTGTPQRR423 pKa = 11.84THH425 pKa = 6.25FLSVFKK431 pKa = 10.83VPKK434 pKa = 10.39GMSLSLGRR442 pKa = 11.84LDD444 pKa = 4.94ALL446 pKa = 4.35
MM1 pKa = 7.46KK2 pKa = 10.25RR3 pKa = 11.84RR4 pKa = 11.84EE5 pKa = 4.1TTEE8 pKa = 3.74QLTSRR13 pKa = 11.84LDD15 pKa = 3.48AVQEE19 pKa = 4.1KK20 pKa = 10.18ILAHH24 pKa = 6.48FEE26 pKa = 4.26TTSTEE31 pKa = 3.84LAAHH35 pKa = 6.34IEE37 pKa = 4.17YY38 pKa = 9.68WSLVRR43 pKa = 11.84QEE45 pKa = 4.91SVILYY50 pKa = 6.66YY51 pKa = 10.67ARR53 pKa = 11.84HH54 pKa = 6.05LGVNRR59 pKa = 11.84LGLQPVPAQAISEE72 pKa = 4.15GRR74 pKa = 11.84AKK76 pKa = 10.16QAIGMQLVLEE86 pKa = 4.27SLQRR90 pKa = 11.84SPYY93 pKa = 10.36ARR95 pKa = 11.84EE96 pKa = 3.39EE97 pKa = 3.82WTLQDD102 pKa = 3.49VSLEE106 pKa = 4.22KK107 pKa = 11.15YY108 pKa = 10.13SVQPKK113 pKa = 8.58NTFKK117 pKa = 11.03KK118 pKa = 10.45DD119 pKa = 3.08GTQVHH124 pKa = 5.8VKK126 pKa = 10.18YY127 pKa = 10.86GPDD130 pKa = 3.3PTDD133 pKa = 3.77FMPYY137 pKa = 9.77VLWGSIYY144 pKa = 10.69YY145 pKa = 10.02QDD147 pKa = 5.82VDD149 pKa = 4.52DD150 pKa = 4.97KK151 pKa = 9.07WHH153 pKa = 6.45KK154 pKa = 11.03VPGQVDD160 pKa = 3.53YY161 pKa = 11.49HH162 pKa = 7.07GLFYY166 pKa = 10.61MEE168 pKa = 4.6GKK170 pKa = 10.12EE171 pKa = 4.03KK172 pKa = 9.89IYY174 pKa = 10.96YY175 pKa = 10.39EE176 pKa = 4.09NFKK179 pKa = 10.88EE180 pKa = 4.0DD181 pKa = 3.86AIKK184 pKa = 10.62FGMPGRR190 pKa = 11.84WIVNVKK196 pKa = 8.4NTAVPSFSRR205 pKa = 11.84QTSSAFAGGRR215 pKa = 11.84PGADD219 pKa = 2.94STDD222 pKa = 3.47APEE225 pKa = 4.61APSTSRR231 pKa = 11.84WSPRR235 pKa = 11.84ASNTTQAQEE244 pKa = 4.34SNQVSGPSTRR254 pKa = 11.84RR255 pKa = 11.84GRR257 pKa = 11.84GRR259 pKa = 11.84RR260 pKa = 11.84SPSPGEE266 pKa = 4.26GPSSSRR272 pKa = 11.84RR273 pKa = 11.84RR274 pKa = 11.84RR275 pKa = 11.84LPTAARR281 pKa = 11.84RR282 pKa = 11.84TPSPEE287 pKa = 3.2RR288 pKa = 11.84HH289 pKa = 5.36SRR291 pKa = 11.84SRR293 pKa = 11.84SRR295 pKa = 11.84DD296 pKa = 2.81WRR298 pKa = 11.84GGRR301 pKa = 11.84GGGGGLRR308 pKa = 11.84GRR310 pKa = 11.84GRR312 pKa = 11.84QGEE315 pKa = 4.44SAGRR319 pKa = 11.84SAGPHH324 pKa = 5.39SRR326 pKa = 11.84SSQSPAPTPPTPGQVGGRR344 pKa = 11.84HH345 pKa = 4.63RR346 pKa = 11.84TAEE349 pKa = 4.22PGASRR354 pKa = 11.84LEE356 pKa = 4.01RR357 pKa = 11.84LIQEE361 pKa = 4.31ARR363 pKa = 11.84DD364 pKa = 3.7PPVVLIRR371 pKa = 11.84GTPNGVKK378 pKa = 10.2CLRR381 pKa = 11.84YY382 pKa = 9.56RR383 pKa = 11.84LRR385 pKa = 11.84TQNQHH390 pKa = 6.65LYY392 pKa = 10.99DD393 pKa = 4.01SVTSTFFWVNKK404 pKa = 9.72NGKK407 pKa = 6.35QQEE410 pKa = 4.23GRR412 pKa = 11.84IMLSFTGTPQRR423 pKa = 11.84THH425 pKa = 6.25FLSVFKK431 pKa = 10.83VPKK434 pKa = 10.39GMSLSLGRR442 pKa = 11.84LDD444 pKa = 4.94ALL446 pKa = 4.35
Molecular weight: 49.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2343 |
97 |
620 |
390.5 |
43.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.719 ± 0.347 | 2.091 ± 0.805 |
6.274 ± 0.649 | 6.018 ± 0.513 |
4.225 ± 0.504 | 7.896 ± 1.159 |
2.006 ± 0.177 | 4.14 ± 0.638 |
4.14 ± 0.851 | 8.792 ± 0.82 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.494 ± 0.31 | 3.286 ± 0.685 |
6.701 ± 0.881 | 4.78 ± 0.586 |
7.085 ± 0.913 | 7.085 ± 0.738 |
6.701 ± 0.518 | 7.128 ± 0.916 |
1.28 ± 0.348 | 3.158 ± 0.371 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
