
Methylovirgula sp. 4M-Z18
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Beijerinckiaceae; Methylovirgula; unclassified Methylovirgula
Average proteome isoelectric point is 6.66
Get precalculated fractions of proteins
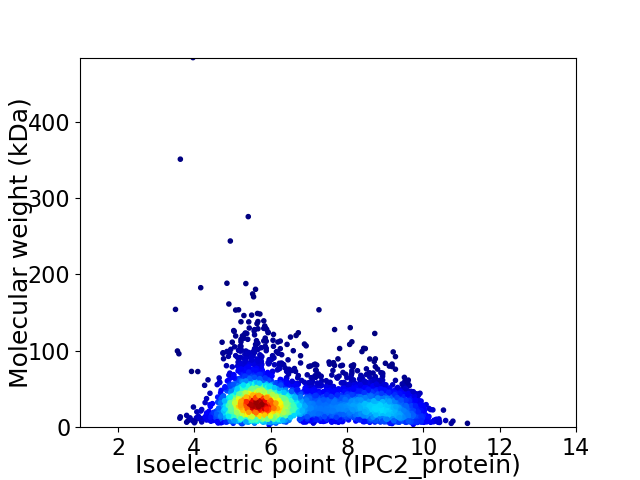
Virtual 2D-PAGE plot for 4535 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A371WU82|A0A371WU82_9RHIZ Exodeoxyribonuclease III OS=Methylovirgula sp. 4M-Z18 OX=2293567 GN=xth PE=3 SV=1
MM1 pKa = 7.25GVLMQINATFGAGSTTNFEE20 pKa = 4.48SAVNSAISFFDD31 pKa = 3.31KK32 pKa = 11.13AFTNNITLNINFNEE46 pKa = 4.0ISQAGGGLSSSSTEE60 pKa = 4.58LINDD64 pKa = 4.68GYY66 pKa = 11.0SAISTALKK74 pKa = 9.47TLNQTADD81 pKa = 3.64QISAAAALPASDD93 pKa = 4.65PFGSTSTASVSSAEE107 pKa = 4.0AKK109 pKa = 10.78ALGLSLNGYY118 pKa = 7.54TGADD122 pKa = 3.29GTVNLNDD129 pKa = 3.67AALPSGDD136 pKa = 2.97SWAWSQNSIGATQFDD151 pKa = 3.91AVGVIEE157 pKa = 4.68HH158 pKa = 7.21EE159 pKa = 4.08ISEE162 pKa = 4.37VMGRR166 pKa = 11.84IAGTTDD172 pKa = 2.54NGAQAGTLLDD182 pKa = 4.08TPEE185 pKa = 3.67AWHH188 pKa = 6.95IYY190 pKa = 8.67GASGSINTGTSASGTYY206 pKa = 10.37LSLNGGTTNLGAIGEE221 pKa = 4.51TGGGDD226 pKa = 3.35LADD229 pKa = 3.37WTDD232 pKa = 4.87PINATNDD239 pKa = 2.91AFGLIKK245 pKa = 10.55AGAVMQVTTTDD256 pKa = 3.36LRR258 pKa = 11.84EE259 pKa = 3.96MSTLGYY265 pKa = 10.48NLASEE270 pKa = 5.28ISQQNYY276 pKa = 8.95DD277 pKa = 3.42ASGKK281 pKa = 10.31LLGATVTVVDD291 pKa = 3.92SQQNVDD297 pKa = 3.31TVQIGFGATATTVTFSDD314 pKa = 3.5PAYY317 pKa = 9.11PNSPQTYY324 pKa = 8.76AAGAGALVLLSSGGTEE340 pKa = 4.1VNISSGSTTNEE351 pKa = 3.82VIISAGSDD359 pKa = 3.35SVTGGSGASLILSLGSGIYY378 pKa = 9.85RR379 pKa = 11.84AGSGTTQFALLSGNGEE395 pKa = 4.15VLASNSSISLGGGWNGFVEE414 pKa = 4.87GSGDD418 pKa = 3.8TITTLGGLGGGRR430 pKa = 11.84PDD432 pKa = 4.9VIGSSNNVTSTNVNDD447 pKa = 4.91FIEE450 pKa = 4.59VGDD453 pKa = 3.94AAGTTSANNVVHH465 pKa = 6.7FNAGGGTLALLDD477 pKa = 3.65NAQANVYY484 pKa = 9.94GDD486 pKa = 3.96HH487 pKa = 6.21LTVYY491 pKa = 10.35VDD493 pKa = 3.52NSSDD497 pKa = 3.5SLALQGSGDD506 pKa = 3.91NVTVDD511 pKa = 3.52TSNVSISVGGNGGAAGVATDD531 pKa = 3.93DD532 pKa = 3.98TVTFDD537 pKa = 5.03ASGTLNVSDD546 pKa = 4.36NSHH549 pKa = 5.22VDD551 pKa = 3.26AYY553 pKa = 10.53GSSVLVTAGTNDD565 pKa = 3.38TVGVQGSNDD574 pKa = 3.12AVHH577 pKa = 7.03ANGNNDD583 pKa = 2.72IVSIGGNGGTFNFSNNDD600 pKa = 3.05GVTVAGTGTTVNEE613 pKa = 4.48LDD615 pKa = 3.74NSHH618 pKa = 6.11VDD620 pKa = 3.15AYY622 pKa = 11.01GNNFAANAGTGDD634 pKa = 3.55TLGVHH639 pKa = 6.77GGGITVNASGNNDD652 pKa = 3.74TVWAGGNSSTFSFADD667 pKa = 3.66DD668 pKa = 3.82DD669 pKa = 4.5TVTFAGTGGTLNEE682 pKa = 4.62SDD684 pKa = 4.14NSHH687 pKa = 6.2VDD689 pKa = 3.26ATGSNFWVNAGTGDD703 pKa = 3.57TVGVHH708 pKa = 6.2GGGITVNASGNNDD721 pKa = 3.74TVWAGGNGQAFSFATDD737 pKa = 3.79DD738 pKa = 4.08SIHH741 pKa = 6.79FYY743 pKa = 9.92GTGSTLNEE751 pKa = 4.3FDD753 pKa = 4.0NSHH756 pKa = 6.56IDD758 pKa = 3.67GFGSNLWVNAGTNDD772 pKa = 3.52TLGVQGSGITANANGNTDD790 pKa = 3.89TVWASGNGQAFSFATDD806 pKa = 3.37DD807 pKa = 3.52SIYY810 pKa = 10.67FHH812 pKa = 6.61GTGGTLNEE820 pKa = 4.49SDD822 pKa = 4.24SSHH825 pKa = 7.17VDD827 pKa = 3.03ASGNGVWVNAGSNDD841 pKa = 3.38TLGLHH846 pKa = 6.91GSAITANASGSNDD859 pKa = 3.79TVWAGGNGQAFSFTTDD875 pKa = 2.97DD876 pKa = 4.3SINLGGSGGVVNEE889 pKa = 4.55FDD891 pKa = 3.79NSHH894 pKa = 6.88VDD896 pKa = 2.91ISGNGLTANVGNNDD910 pKa = 2.94TFGAHH915 pKa = 6.28GSSDD919 pKa = 3.58VVHH922 pKa = 7.06ANGNSDD928 pKa = 3.94TVWLGGNGQNASAHH942 pKa = 6.01DD943 pKa = 3.78AVYY946 pKa = 9.59FAGSAGVLNLADD958 pKa = 4.12NSTVDD963 pKa = 3.29ATGNNVWANLGNNDD977 pKa = 3.77TLNLFGSGDD986 pKa = 3.74GFSASGSGDD995 pKa = 3.78VVWLNGVRR1003 pKa = 11.84VRR1005 pKa = 4.8
MM1 pKa = 7.25GVLMQINATFGAGSTTNFEE20 pKa = 4.48SAVNSAISFFDD31 pKa = 3.31KK32 pKa = 11.13AFTNNITLNINFNEE46 pKa = 4.0ISQAGGGLSSSSTEE60 pKa = 4.58LINDD64 pKa = 4.68GYY66 pKa = 11.0SAISTALKK74 pKa = 9.47TLNQTADD81 pKa = 3.64QISAAAALPASDD93 pKa = 4.65PFGSTSTASVSSAEE107 pKa = 4.0AKK109 pKa = 10.78ALGLSLNGYY118 pKa = 7.54TGADD122 pKa = 3.29GTVNLNDD129 pKa = 3.67AALPSGDD136 pKa = 2.97SWAWSQNSIGATQFDD151 pKa = 3.91AVGVIEE157 pKa = 4.68HH158 pKa = 7.21EE159 pKa = 4.08ISEE162 pKa = 4.37VMGRR166 pKa = 11.84IAGTTDD172 pKa = 2.54NGAQAGTLLDD182 pKa = 4.08TPEE185 pKa = 3.67AWHH188 pKa = 6.95IYY190 pKa = 8.67GASGSINTGTSASGTYY206 pKa = 10.37LSLNGGTTNLGAIGEE221 pKa = 4.51TGGGDD226 pKa = 3.35LADD229 pKa = 3.37WTDD232 pKa = 4.87PINATNDD239 pKa = 2.91AFGLIKK245 pKa = 10.55AGAVMQVTTTDD256 pKa = 3.36LRR258 pKa = 11.84EE259 pKa = 3.96MSTLGYY265 pKa = 10.48NLASEE270 pKa = 5.28ISQQNYY276 pKa = 8.95DD277 pKa = 3.42ASGKK281 pKa = 10.31LLGATVTVVDD291 pKa = 3.92SQQNVDD297 pKa = 3.31TVQIGFGATATTVTFSDD314 pKa = 3.5PAYY317 pKa = 9.11PNSPQTYY324 pKa = 8.76AAGAGALVLLSSGGTEE340 pKa = 4.1VNISSGSTTNEE351 pKa = 3.82VIISAGSDD359 pKa = 3.35SVTGGSGASLILSLGSGIYY378 pKa = 9.85RR379 pKa = 11.84AGSGTTQFALLSGNGEE395 pKa = 4.15VLASNSSISLGGGWNGFVEE414 pKa = 4.87GSGDD418 pKa = 3.8TITTLGGLGGGRR430 pKa = 11.84PDD432 pKa = 4.9VIGSSNNVTSTNVNDD447 pKa = 4.91FIEE450 pKa = 4.59VGDD453 pKa = 3.94AAGTTSANNVVHH465 pKa = 6.7FNAGGGTLALLDD477 pKa = 3.65NAQANVYY484 pKa = 9.94GDD486 pKa = 3.96HH487 pKa = 6.21LTVYY491 pKa = 10.35VDD493 pKa = 3.52NSSDD497 pKa = 3.5SLALQGSGDD506 pKa = 3.91NVTVDD511 pKa = 3.52TSNVSISVGGNGGAAGVATDD531 pKa = 3.93DD532 pKa = 3.98TVTFDD537 pKa = 5.03ASGTLNVSDD546 pKa = 4.36NSHH549 pKa = 5.22VDD551 pKa = 3.26AYY553 pKa = 10.53GSSVLVTAGTNDD565 pKa = 3.38TVGVQGSNDD574 pKa = 3.12AVHH577 pKa = 7.03ANGNNDD583 pKa = 2.72IVSIGGNGGTFNFSNNDD600 pKa = 3.05GVTVAGTGTTVNEE613 pKa = 4.48LDD615 pKa = 3.74NSHH618 pKa = 6.11VDD620 pKa = 3.15AYY622 pKa = 11.01GNNFAANAGTGDD634 pKa = 3.55TLGVHH639 pKa = 6.77GGGITVNASGNNDD652 pKa = 3.74TVWAGGNSSTFSFADD667 pKa = 3.66DD668 pKa = 3.82DD669 pKa = 4.5TVTFAGTGGTLNEE682 pKa = 4.62SDD684 pKa = 4.14NSHH687 pKa = 6.2VDD689 pKa = 3.26ATGSNFWVNAGTGDD703 pKa = 3.57TVGVHH708 pKa = 6.2GGGITVNASGNNDD721 pKa = 3.74TVWAGGNGQAFSFATDD737 pKa = 3.79DD738 pKa = 4.08SIHH741 pKa = 6.79FYY743 pKa = 9.92GTGSTLNEE751 pKa = 4.3FDD753 pKa = 4.0NSHH756 pKa = 6.56IDD758 pKa = 3.67GFGSNLWVNAGTNDD772 pKa = 3.52TLGVQGSGITANANGNTDD790 pKa = 3.89TVWASGNGQAFSFATDD806 pKa = 3.37DD807 pKa = 3.52SIYY810 pKa = 10.67FHH812 pKa = 6.61GTGGTLNEE820 pKa = 4.49SDD822 pKa = 4.24SSHH825 pKa = 7.17VDD827 pKa = 3.03ASGNGVWVNAGSNDD841 pKa = 3.38TLGLHH846 pKa = 6.91GSAITANASGSNDD859 pKa = 3.79TVWAGGNGQAFSFTTDD875 pKa = 2.97DD876 pKa = 4.3SINLGGSGGVVNEE889 pKa = 4.55FDD891 pKa = 3.79NSHH894 pKa = 6.88VDD896 pKa = 2.91ISGNGLTANVGNNDD910 pKa = 2.94TFGAHH915 pKa = 6.28GSSDD919 pKa = 3.58VVHH922 pKa = 7.06ANGNSDD928 pKa = 3.94TVWLGGNGQNASAHH942 pKa = 6.01DD943 pKa = 3.78AVYY946 pKa = 9.59FAGSAGVLNLADD958 pKa = 4.12NSTVDD963 pKa = 3.29ATGNNVWANLGNNDD977 pKa = 3.77TLNLFGSGDD986 pKa = 3.74GFSASGSGDD995 pKa = 3.78VVWLNGVRR1003 pKa = 11.84VRR1005 pKa = 4.8
Molecular weight: 99.75 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A371WPG6|A0A371WPG6_9RHIZ Arylesterase OS=Methylovirgula sp. 4M-Z18 OX=2293567 GN=DYH55_14740 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84AVIAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84AVIAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
Molecular weight: 5.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1443161 |
28 |
5431 |
318.2 |
34.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.56 ± 0.049 | 0.906 ± 0.013 |
5.631 ± 0.031 | 5.142 ± 0.04 |
3.836 ± 0.027 | 8.436 ± 0.098 |
2.27 ± 0.018 | 5.416 ± 0.026 |
3.569 ± 0.035 | 10.121 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.43 ± 0.018 | 2.802 ± 0.033 |
5.138 ± 0.031 | 3.346 ± 0.022 |
6.609 ± 0.054 | 5.464 ± 0.048 |
5.346 ± 0.035 | 7.331 ± 0.026 |
1.34 ± 0.014 | 2.31 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
