
Flavobacteriaceae bacterium F202Z8
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; unclassified Flavobacteriaceae
Average proteome isoelectric point is 6.33
Get precalculated fractions of proteins
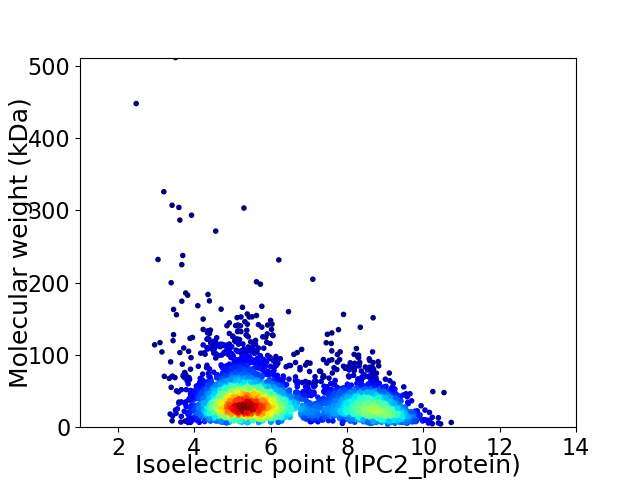
Virtual 2D-PAGE plot for 3847 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5B7SYN8|A0A5B7SYN8_9FLAO Response regulator transcription factor OS=Flavobacteriaceae bacterium F202Z8 OX=2583587 GN=FGM00_08155 PE=4 SV=1
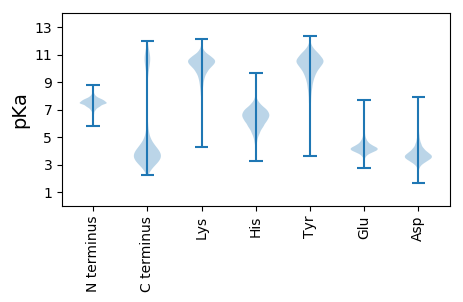
MM1 pKa = 7.7LSGALRR7 pKa = 11.84KK8 pKa = 10.06YY9 pKa = 10.62GLSFLLGACLVFMLLLASSCDD30 pKa = 3.7NEE32 pKa = 5.08DD33 pKa = 4.08VLDD36 pKa = 4.47LDD38 pKa = 4.19EE39 pKa = 4.5TAMTNDD45 pKa = 5.4DD46 pKa = 4.66IVTCTDD52 pKa = 3.75GIMNGDD58 pKa = 3.68EE59 pKa = 4.08TGIDD63 pKa = 3.54CGGACTPCEE72 pKa = 4.0AGEE75 pKa = 4.06EE76 pKa = 4.04LGRR79 pKa = 11.84RR80 pKa = 11.84AEE82 pKa = 4.69LYY84 pKa = 8.43VTNNANGDD92 pKa = 3.15ISRR95 pKa = 11.84YY96 pKa = 9.82SVTGDD101 pKa = 3.04SLTTFTTASTAAEE114 pKa = 4.59GIYY117 pKa = 10.34YY118 pKa = 10.2SSKK121 pKa = 11.17DD122 pKa = 3.46GVLVQASRR130 pKa = 11.84SGLQLDD136 pKa = 4.77AYY138 pKa = 10.67SGIAGIMEE146 pKa = 5.06DD147 pKa = 3.74SALDD151 pKa = 3.52ASFSSTTDD159 pKa = 3.18LEE161 pKa = 4.65SPRR164 pKa = 11.84EE165 pKa = 3.88LAVNGDD171 pKa = 3.99MYY173 pKa = 11.42VVSDD177 pKa = 3.4NGSNKK182 pKa = 9.43FFVYY186 pKa = 9.63TKK188 pKa = 10.63NGDD191 pKa = 3.99SFSLTSTVEE200 pKa = 3.5IPFPVWGITFRR211 pKa = 11.84GSDD214 pKa = 3.84LYY216 pKa = 11.45AVVDD220 pKa = 3.94NSSDD224 pKa = 3.39LAVFNNFQASISAPVCALNPSKK246 pKa = 10.57RR247 pKa = 11.84VVIEE251 pKa = 4.47GIVRR255 pKa = 11.84THH257 pKa = 5.78GLVYY261 pKa = 10.66NAADD265 pKa = 5.25DD266 pKa = 3.85IMIMTDD272 pKa = 3.02IGDD275 pKa = 4.14AANTTDD281 pKa = 4.76DD282 pKa = 3.99GAFHH286 pKa = 6.81VISEE290 pKa = 4.51FAATFDD296 pKa = 4.19LLSDD300 pKa = 3.91GEE302 pKa = 4.62VYY304 pKa = 10.39PLSRR308 pKa = 11.84QVRR311 pKa = 11.84VAGPSTMMGNPIDD324 pKa = 3.64VAYY327 pKa = 10.39DD328 pKa = 3.47SEE330 pKa = 4.45EE331 pKa = 3.84NAVYY335 pKa = 10.19ISEE338 pKa = 4.2VGNGKK343 pKa = 9.25VLGFTAQGGGDD354 pKa = 3.43RR355 pKa = 11.84APWFVKK361 pKa = 10.28DD362 pKa = 4.32LPAASSIYY370 pKa = 10.13FSSDD374 pKa = 2.82EE375 pKa = 4.15TDD377 pKa = 3.54DD378 pKa = 4.77DD379 pKa = 4.53LGEE382 pKa = 3.85EE383 pKa = 4.06AMAGNTVLYY392 pKa = 8.44ATSTANGNIYY402 pKa = 10.65VYY404 pKa = 10.98DD405 pKa = 3.73AMGNALKK412 pKa = 9.61TVTSQSEE419 pKa = 4.26SSEE422 pKa = 4.1GIYY425 pKa = 8.85YY426 pKa = 8.18TALDD430 pKa = 5.29DD431 pKa = 4.69ILVQASRR438 pKa = 11.84SALQLEE444 pKa = 5.62GYY446 pKa = 10.7AEE448 pKa = 4.84FSTVPDD454 pKa = 3.97GAIGAASFTSSKK466 pKa = 11.29DD467 pKa = 3.23LMSPRR472 pKa = 11.84EE473 pKa = 3.64IAVYY477 pKa = 10.2GNKK480 pKa = 9.53IVVADD485 pKa = 3.52NAEE488 pKa = 4.23NIFYY492 pKa = 10.59VYY494 pKa = 10.37SHH496 pKa = 7.21DD497 pKa = 3.67MFSFSLINTFRR508 pKa = 11.84FEE510 pKa = 4.58CRR512 pKa = 11.84GIWGITFKK520 pKa = 11.18GDD522 pKa = 3.37DD523 pKa = 3.96LLAVVDD529 pKa = 4.03NTGDD533 pKa = 3.55LAIFSDD539 pKa = 4.63FFTNYY544 pKa = 9.08TVDD547 pKa = 3.81GNITPEE553 pKa = 4.49KK554 pKa = 9.97IITIQGIVRR563 pKa = 11.84THH565 pKa = 6.85GIDD568 pKa = 3.34YY569 pKa = 10.78SAADD573 pKa = 4.69DD574 pKa = 4.54LLVMTDD580 pKa = 3.89IGDD583 pKa = 4.06AANGTDD589 pKa = 4.38DD590 pKa = 5.69GGFQLIQEE598 pKa = 4.74FSAKK602 pKa = 10.05LDD604 pKa = 4.57AISNGGTLVLSDD616 pKa = 3.53QTRR619 pKa = 11.84VSGPGTMMGNPIDD632 pKa = 3.51IAYY635 pKa = 10.42DD636 pKa = 3.59NDD638 pKa = 3.75SKK640 pKa = 10.11TVYY643 pKa = 9.82IAEE646 pKa = 4.33IGNGKK651 pKa = 9.05ILGFSDD657 pKa = 3.52VLNASGDD664 pKa = 3.71VAPTFNWDD672 pKa = 3.21LNQASSIFLYY682 pKa = 11.12NNN684 pKa = 3.13
MM1 pKa = 7.7LSGALRR7 pKa = 11.84KK8 pKa = 10.06YY9 pKa = 10.62GLSFLLGACLVFMLLLASSCDD30 pKa = 3.7NEE32 pKa = 5.08DD33 pKa = 4.08VLDD36 pKa = 4.47LDD38 pKa = 4.19EE39 pKa = 4.5TAMTNDD45 pKa = 5.4DD46 pKa = 4.66IVTCTDD52 pKa = 3.75GIMNGDD58 pKa = 3.68EE59 pKa = 4.08TGIDD63 pKa = 3.54CGGACTPCEE72 pKa = 4.0AGEE75 pKa = 4.06EE76 pKa = 4.04LGRR79 pKa = 11.84RR80 pKa = 11.84AEE82 pKa = 4.69LYY84 pKa = 8.43VTNNANGDD92 pKa = 3.15ISRR95 pKa = 11.84YY96 pKa = 9.82SVTGDD101 pKa = 3.04SLTTFTTASTAAEE114 pKa = 4.59GIYY117 pKa = 10.34YY118 pKa = 10.2SSKK121 pKa = 11.17DD122 pKa = 3.46GVLVQASRR130 pKa = 11.84SGLQLDD136 pKa = 4.77AYY138 pKa = 10.67SGIAGIMEE146 pKa = 5.06DD147 pKa = 3.74SALDD151 pKa = 3.52ASFSSTTDD159 pKa = 3.18LEE161 pKa = 4.65SPRR164 pKa = 11.84EE165 pKa = 3.88LAVNGDD171 pKa = 3.99MYY173 pKa = 11.42VVSDD177 pKa = 3.4NGSNKK182 pKa = 9.43FFVYY186 pKa = 9.63TKK188 pKa = 10.63NGDD191 pKa = 3.99SFSLTSTVEE200 pKa = 3.5IPFPVWGITFRR211 pKa = 11.84GSDD214 pKa = 3.84LYY216 pKa = 11.45AVVDD220 pKa = 3.94NSSDD224 pKa = 3.39LAVFNNFQASISAPVCALNPSKK246 pKa = 10.57RR247 pKa = 11.84VVIEE251 pKa = 4.47GIVRR255 pKa = 11.84THH257 pKa = 5.78GLVYY261 pKa = 10.66NAADD265 pKa = 5.25DD266 pKa = 3.85IMIMTDD272 pKa = 3.02IGDD275 pKa = 4.14AANTTDD281 pKa = 4.76DD282 pKa = 3.99GAFHH286 pKa = 6.81VISEE290 pKa = 4.51FAATFDD296 pKa = 4.19LLSDD300 pKa = 3.91GEE302 pKa = 4.62VYY304 pKa = 10.39PLSRR308 pKa = 11.84QVRR311 pKa = 11.84VAGPSTMMGNPIDD324 pKa = 3.64VAYY327 pKa = 10.39DD328 pKa = 3.47SEE330 pKa = 4.45EE331 pKa = 3.84NAVYY335 pKa = 10.19ISEE338 pKa = 4.2VGNGKK343 pKa = 9.25VLGFTAQGGGDD354 pKa = 3.43RR355 pKa = 11.84APWFVKK361 pKa = 10.28DD362 pKa = 4.32LPAASSIYY370 pKa = 10.13FSSDD374 pKa = 2.82EE375 pKa = 4.15TDD377 pKa = 3.54DD378 pKa = 4.77DD379 pKa = 4.53LGEE382 pKa = 3.85EE383 pKa = 4.06AMAGNTVLYY392 pKa = 8.44ATSTANGNIYY402 pKa = 10.65VYY404 pKa = 10.98DD405 pKa = 3.73AMGNALKK412 pKa = 9.61TVTSQSEE419 pKa = 4.26SSEE422 pKa = 4.1GIYY425 pKa = 8.85YY426 pKa = 8.18TALDD430 pKa = 5.29DD431 pKa = 4.69ILVQASRR438 pKa = 11.84SALQLEE444 pKa = 5.62GYY446 pKa = 10.7AEE448 pKa = 4.84FSTVPDD454 pKa = 3.97GAIGAASFTSSKK466 pKa = 11.29DD467 pKa = 3.23LMSPRR472 pKa = 11.84EE473 pKa = 3.64IAVYY477 pKa = 10.2GNKK480 pKa = 9.53IVVADD485 pKa = 3.52NAEE488 pKa = 4.23NIFYY492 pKa = 10.59VYY494 pKa = 10.37SHH496 pKa = 7.21DD497 pKa = 3.67MFSFSLINTFRR508 pKa = 11.84FEE510 pKa = 4.58CRR512 pKa = 11.84GIWGITFKK520 pKa = 11.18GDD522 pKa = 3.37DD523 pKa = 3.96LLAVVDD529 pKa = 4.03NTGDD533 pKa = 3.55LAIFSDD539 pKa = 4.63FFTNYY544 pKa = 9.08TVDD547 pKa = 3.81GNITPEE553 pKa = 4.49KK554 pKa = 9.97IITIQGIVRR563 pKa = 11.84THH565 pKa = 6.85GIDD568 pKa = 3.34YY569 pKa = 10.78SAADD573 pKa = 4.69DD574 pKa = 4.54LLVMTDD580 pKa = 3.89IGDD583 pKa = 4.06AANGTDD589 pKa = 4.38DD590 pKa = 5.69GGFQLIQEE598 pKa = 4.74FSAKK602 pKa = 10.05LDD604 pKa = 4.57AISNGGTLVLSDD616 pKa = 3.53QTRR619 pKa = 11.84VSGPGTMMGNPIDD632 pKa = 3.51IAYY635 pKa = 10.42DD636 pKa = 3.59NDD638 pKa = 3.75SKK640 pKa = 10.11TVYY643 pKa = 9.82IAEE646 pKa = 4.33IGNGKK651 pKa = 9.05ILGFSDD657 pKa = 3.52VLNASGDD664 pKa = 3.71VAPTFNWDD672 pKa = 3.21LNQASSIFLYY682 pKa = 11.12NNN684 pKa = 3.13
Molecular weight: 72.83 kDa
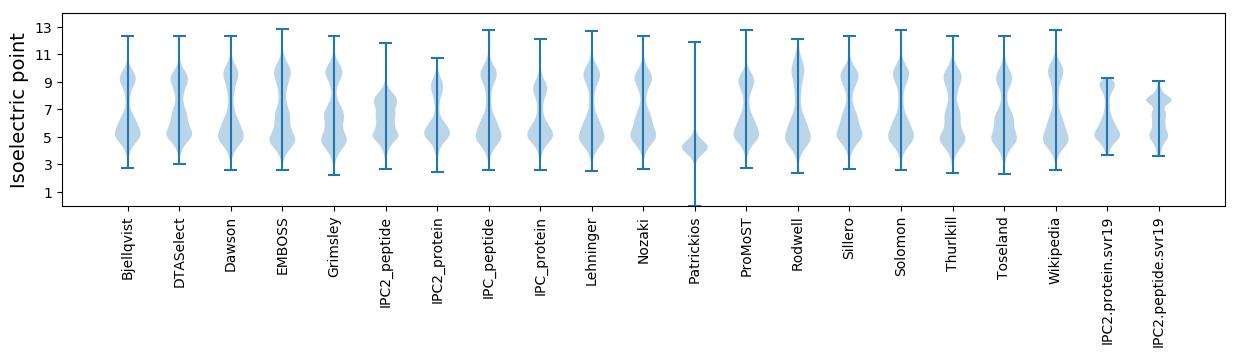
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5B7SPQ6|A0A5B7SPQ6_9FLAO Acyl-CoA carboxylase subunit beta OS=Flavobacteriaceae bacterium F202Z8 OX=2583587 GN=FGM00_08650 PE=4 SV=1
MM1 pKa = 7.67RR2 pKa = 11.84KK3 pKa = 9.86AEE5 pKa = 3.98NRR7 pKa = 11.84EE8 pKa = 3.58QRR10 pKa = 11.84MEE12 pKa = 4.01NGEE15 pKa = 3.83KK16 pKa = 10.14RR17 pKa = 11.84KK18 pKa = 9.81EE19 pKa = 3.81KK20 pKa = 10.74RR21 pKa = 11.84EE22 pKa = 3.6WRR24 pKa = 11.84MEE26 pKa = 3.76NGEE29 pKa = 3.87WRR31 pKa = 11.84KK32 pKa = 9.42FRR34 pKa = 11.84VKK36 pKa = 10.54NSNPNRR42 pKa = 11.84YY43 pKa = 6.38TLRR46 pKa = 11.84LFLVSCWRR54 pKa = 11.84MVCSRR59 pKa = 11.84IKK61 pKa = 10.6FKK63 pKa = 10.99LLSKK67 pKa = 10.91GG68 pKa = 3.37
MM1 pKa = 7.67RR2 pKa = 11.84KK3 pKa = 9.86AEE5 pKa = 3.98NRR7 pKa = 11.84EE8 pKa = 3.58QRR10 pKa = 11.84MEE12 pKa = 4.01NGEE15 pKa = 3.83KK16 pKa = 10.14RR17 pKa = 11.84KK18 pKa = 9.81EE19 pKa = 3.81KK20 pKa = 10.74RR21 pKa = 11.84EE22 pKa = 3.6WRR24 pKa = 11.84MEE26 pKa = 3.76NGEE29 pKa = 3.87WRR31 pKa = 11.84KK32 pKa = 9.42FRR34 pKa = 11.84VKK36 pKa = 10.54NSNPNRR42 pKa = 11.84YY43 pKa = 6.38TLRR46 pKa = 11.84LFLVSCWRR54 pKa = 11.84MVCSRR59 pKa = 11.84IKK61 pKa = 10.6FKK63 pKa = 10.99LLSKK67 pKa = 10.91GG68 pKa = 3.37
Molecular weight: 8.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1374296 |
38 |
4852 |
357.2 |
40.07 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.062 ± 0.035 | 0.745 ± 0.014 |
5.978 ± 0.053 | 6.692 ± 0.037 |
5.043 ± 0.032 | 6.99 ± 0.048 |
1.781 ± 0.021 | 7.124 ± 0.032 |
6.858 ± 0.065 | 9.306 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.285 ± 0.023 | 5.488 ± 0.04 |
3.647 ± 0.026 | 3.38 ± 0.022 |
3.919 ± 0.03 | 6.335 ± 0.032 |
5.953 ± 0.059 | 6.418 ± 0.038 |
1.14 ± 0.014 | 3.855 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
