
Cloacimonas acidaminovorans (strain Evry)
Taxonomy: cellular organisms; Bacteria; FCB group; Candidatus Cloacimonetes; Candidatus Cloacimonas; Candidatus Cloacimonas acidaminovorans
Average proteome isoelectric point is 6.56
Get precalculated fractions of proteins
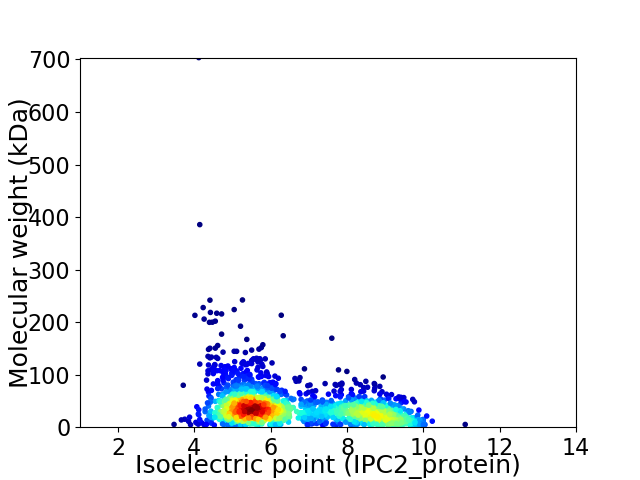
Virtual 2D-PAGE plot for 1813 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
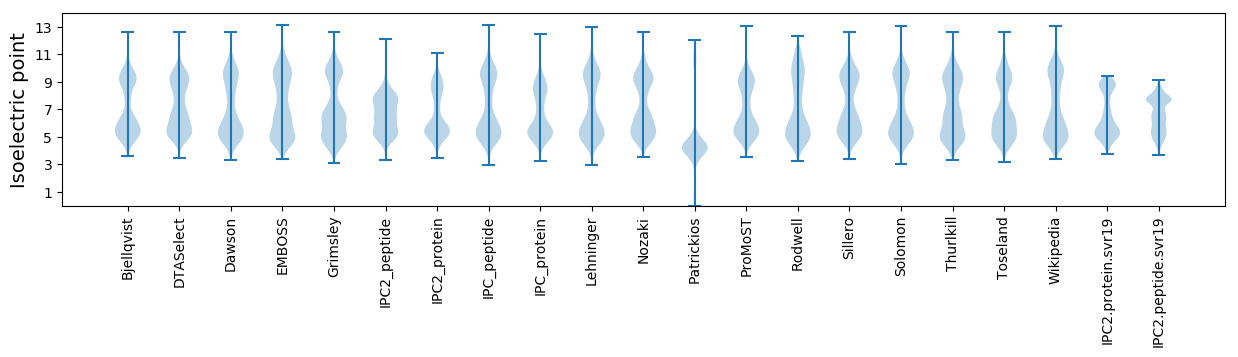
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B0VJF9|B0VJF9_CLOAI Diaminopimelate decarboxylase (DAP decarboxylase) OS=Cloacimonas acidaminovorans (strain Evry) OX=459349 GN=lysA PE=4 SV=1
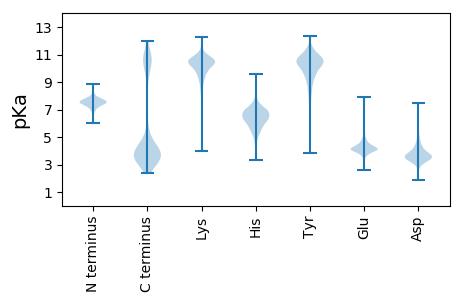
MM1 pKa = 7.2KK2 pKa = 9.26RR3 pKa = 11.84TYY5 pKa = 10.51IFIGILFLALCLNAQPWEE23 pKa = 4.0MDD25 pKa = 2.81NSVFNPSGVPSFSFSQPRR43 pKa = 11.84FMDD46 pKa = 4.31LDD48 pKa = 3.72SDD50 pKa = 4.08GDD52 pKa = 3.65FDD54 pKa = 4.4FWLGNTNRR62 pKa = 11.84PPLYY66 pKa = 9.99LQNNGTATNPVFAPGPDD83 pKa = 3.69LLSGIPYY90 pKa = 9.58LASEE94 pKa = 4.52IAVSADD100 pKa = 3.3MNGDD104 pKa = 4.33GILDD108 pKa = 4.18LVTGGFSGLHH118 pKa = 5.97LFLNSGTNANPVFTEE133 pKa = 3.65SGGYY137 pKa = 9.43FSGLNVGLNPVPDD150 pKa = 3.81VADD153 pKa = 3.38VDD155 pKa = 3.95NDD157 pKa = 3.54GDD159 pKa = 4.2LDD161 pKa = 4.2LVVGLSEE168 pKa = 4.9DD169 pKa = 3.65GSVIIYY175 pKa = 9.71FNTGSAAAGNFSEE188 pKa = 5.42NNCQLLGDD196 pKa = 4.08IGLYY200 pKa = 10.1AYY202 pKa = 9.49PVFCDD207 pKa = 3.91FDD209 pKa = 5.27SDD211 pKa = 3.86GKK213 pKa = 10.86QDD215 pKa = 3.57ILCGRR220 pKa = 11.84DD221 pKa = 2.9AFGFVYY227 pKa = 10.0YY228 pKa = 10.24QNIGTATNPVWEE240 pKa = 4.45EE241 pKa = 3.9NNTLFAGLGMSTYY254 pKa = 9.68WNSPDD259 pKa = 5.29LVDD262 pKa = 5.35LNGDD266 pKa = 3.64GLYY269 pKa = 11.05DD270 pKa = 3.96LVYY273 pKa = 9.45GTAAGPLQYY282 pKa = 10.28YY283 pKa = 7.13VHH285 pKa = 6.98NGTAEE290 pKa = 4.04NPSWQQNTSLFGGVLDD306 pKa = 4.75VGGASNPVFYY316 pKa = 10.95DD317 pKa = 3.46FDD319 pKa = 6.33GDD321 pKa = 3.76GDD323 pKa = 4.69LDD325 pKa = 4.86LISGNQLGYY334 pKa = 11.01VKK336 pKa = 10.15FYY338 pKa = 10.96RR339 pKa = 11.84NTGTAYY345 pKa = 10.41APAWQEE351 pKa = 3.98DD352 pKa = 3.22NSYY355 pKa = 10.38FANIHH360 pKa = 5.71HH361 pKa = 7.09SIYY364 pKa = 10.71SAVTVGDD371 pKa = 3.43VDD373 pKa = 5.07ADD375 pKa = 3.82GLPDD379 pKa = 4.86VILGDD384 pKa = 4.26LNGGLYY390 pKa = 10.25FYY392 pKa = 10.7HH393 pKa = 5.96NTGTGLIEE401 pKa = 4.02QSGVLPAVSVGGWSCPRR418 pKa = 11.84LIDD421 pKa = 3.65MDD423 pKa = 4.15FDD425 pKa = 5.43GDD427 pKa = 3.75LDD429 pKa = 3.97LVVGNEE435 pKa = 3.86AGNLFYY441 pKa = 10.84YY442 pKa = 10.09QNNGTPYY449 pKa = 9.46SPLWEE454 pKa = 4.07LVNGFFGAIDD464 pKa = 3.65VGSDD468 pKa = 3.69CVPTFADD475 pKa = 3.35VDD477 pKa = 3.93EE478 pKa = 6.36DD479 pKa = 4.9DD480 pKa = 5.28DD481 pKa = 6.69LDD483 pKa = 3.95MVTGNLFGEE492 pKa = 4.71VQCFLRR498 pKa = 11.84QRR500 pKa = 11.84QLWVEE505 pKa = 3.95NTTLFSGIEE514 pKa = 4.04TDD516 pKa = 4.06QNAAPALVDD525 pKa = 4.47LDD527 pKa = 3.99HH528 pKa = 7.58DD529 pKa = 4.59GDD531 pKa = 4.07FDD533 pKa = 5.57LVLGDD538 pKa = 3.69YY539 pKa = 11.13DD540 pKa = 3.77GTFKK544 pKa = 10.86FFRR547 pKa = 11.84NLKK550 pKa = 9.83YY551 pKa = 10.51SGSVLNPPQNPIFIIDD567 pKa = 3.92GEE569 pKa = 4.71VTVTWEE575 pKa = 3.95EE576 pKa = 3.92PLIGSTSPFEE586 pKa = 4.09HH587 pKa = 6.35YY588 pKa = 10.23KK589 pKa = 10.26IYY591 pKa = 10.64LDD593 pKa = 4.37GVFCGSTTEE602 pKa = 4.36NIWVFSGLDD611 pKa = 3.06TGVAYY616 pKa = 10.31LVSITAQYY624 pKa = 10.08VAGEE628 pKa = 4.26SLPVTLEE635 pKa = 3.99FIITGNDD642 pKa = 3.43DD643 pKa = 4.3QIVQPITLTNYY654 pKa = 7.75PNPFNPSTTISFTIPAGSKK673 pKa = 8.91GTLEE677 pKa = 3.85IFNVKK682 pKa = 8.03GQKK685 pKa = 9.28VRR687 pKa = 11.84VWNYY691 pKa = 10.28LPSGEE696 pKa = 4.4HH697 pKa = 6.1KK698 pKa = 10.55IVFNGCNEE706 pKa = 3.9QGLPLPSGVYY716 pKa = 9.53FGIMKK721 pKa = 7.74TQNRR725 pKa = 11.84VQIRR729 pKa = 11.84KK730 pKa = 7.91MALVKK735 pKa = 10.76
MM1 pKa = 7.2KK2 pKa = 9.26RR3 pKa = 11.84TYY5 pKa = 10.51IFIGILFLALCLNAQPWEE23 pKa = 4.0MDD25 pKa = 2.81NSVFNPSGVPSFSFSQPRR43 pKa = 11.84FMDD46 pKa = 4.31LDD48 pKa = 3.72SDD50 pKa = 4.08GDD52 pKa = 3.65FDD54 pKa = 4.4FWLGNTNRR62 pKa = 11.84PPLYY66 pKa = 9.99LQNNGTATNPVFAPGPDD83 pKa = 3.69LLSGIPYY90 pKa = 9.58LASEE94 pKa = 4.52IAVSADD100 pKa = 3.3MNGDD104 pKa = 4.33GILDD108 pKa = 4.18LVTGGFSGLHH118 pKa = 5.97LFLNSGTNANPVFTEE133 pKa = 3.65SGGYY137 pKa = 9.43FSGLNVGLNPVPDD150 pKa = 3.81VADD153 pKa = 3.38VDD155 pKa = 3.95NDD157 pKa = 3.54GDD159 pKa = 4.2LDD161 pKa = 4.2LVVGLSEE168 pKa = 4.9DD169 pKa = 3.65GSVIIYY175 pKa = 9.71FNTGSAAAGNFSEE188 pKa = 5.42NNCQLLGDD196 pKa = 4.08IGLYY200 pKa = 10.1AYY202 pKa = 9.49PVFCDD207 pKa = 3.91FDD209 pKa = 5.27SDD211 pKa = 3.86GKK213 pKa = 10.86QDD215 pKa = 3.57ILCGRR220 pKa = 11.84DD221 pKa = 2.9AFGFVYY227 pKa = 10.0YY228 pKa = 10.24QNIGTATNPVWEE240 pKa = 4.45EE241 pKa = 3.9NNTLFAGLGMSTYY254 pKa = 9.68WNSPDD259 pKa = 5.29LVDD262 pKa = 5.35LNGDD266 pKa = 3.64GLYY269 pKa = 11.05DD270 pKa = 3.96LVYY273 pKa = 9.45GTAAGPLQYY282 pKa = 10.28YY283 pKa = 7.13VHH285 pKa = 6.98NGTAEE290 pKa = 4.04NPSWQQNTSLFGGVLDD306 pKa = 4.75VGGASNPVFYY316 pKa = 10.95DD317 pKa = 3.46FDD319 pKa = 6.33GDD321 pKa = 3.76GDD323 pKa = 4.69LDD325 pKa = 4.86LISGNQLGYY334 pKa = 11.01VKK336 pKa = 10.15FYY338 pKa = 10.96RR339 pKa = 11.84NTGTAYY345 pKa = 10.41APAWQEE351 pKa = 3.98DD352 pKa = 3.22NSYY355 pKa = 10.38FANIHH360 pKa = 5.71HH361 pKa = 7.09SIYY364 pKa = 10.71SAVTVGDD371 pKa = 3.43VDD373 pKa = 5.07ADD375 pKa = 3.82GLPDD379 pKa = 4.86VILGDD384 pKa = 4.26LNGGLYY390 pKa = 10.25FYY392 pKa = 10.7HH393 pKa = 5.96NTGTGLIEE401 pKa = 4.02QSGVLPAVSVGGWSCPRR418 pKa = 11.84LIDD421 pKa = 3.65MDD423 pKa = 4.15FDD425 pKa = 5.43GDD427 pKa = 3.75LDD429 pKa = 3.97LVVGNEE435 pKa = 3.86AGNLFYY441 pKa = 10.84YY442 pKa = 10.09QNNGTPYY449 pKa = 9.46SPLWEE454 pKa = 4.07LVNGFFGAIDD464 pKa = 3.65VGSDD468 pKa = 3.69CVPTFADD475 pKa = 3.35VDD477 pKa = 3.93EE478 pKa = 6.36DD479 pKa = 4.9DD480 pKa = 5.28DD481 pKa = 6.69LDD483 pKa = 3.95MVTGNLFGEE492 pKa = 4.71VQCFLRR498 pKa = 11.84QRR500 pKa = 11.84QLWVEE505 pKa = 3.95NTTLFSGIEE514 pKa = 4.04TDD516 pKa = 4.06QNAAPALVDD525 pKa = 4.47LDD527 pKa = 3.99HH528 pKa = 7.58DD529 pKa = 4.59GDD531 pKa = 4.07FDD533 pKa = 5.57LVLGDD538 pKa = 3.69YY539 pKa = 11.13DD540 pKa = 3.77GTFKK544 pKa = 10.86FFRR547 pKa = 11.84NLKK550 pKa = 9.83YY551 pKa = 10.51SGSVLNPPQNPIFIIDD567 pKa = 3.92GEE569 pKa = 4.71VTVTWEE575 pKa = 3.95EE576 pKa = 3.92PLIGSTSPFEE586 pKa = 4.09HH587 pKa = 6.35YY588 pKa = 10.23KK589 pKa = 10.26IYY591 pKa = 10.64LDD593 pKa = 4.37GVFCGSTTEE602 pKa = 4.36NIWVFSGLDD611 pKa = 3.06TGVAYY616 pKa = 10.31LVSITAQYY624 pKa = 10.08VAGEE628 pKa = 4.26SLPVTLEE635 pKa = 3.99FIITGNDD642 pKa = 3.43DD643 pKa = 4.3QIVQPITLTNYY654 pKa = 7.75PNPFNPSTTISFTIPAGSKK673 pKa = 8.91GTLEE677 pKa = 3.85IFNVKK682 pKa = 8.03GQKK685 pKa = 9.28VRR687 pKa = 11.84VWNYY691 pKa = 10.28LPSGEE696 pKa = 4.4HH697 pKa = 6.1KK698 pKa = 10.55IVFNGCNEE706 pKa = 3.9QGLPLPSGVYY716 pKa = 9.53FGIMKK721 pKa = 7.74TQNRR725 pKa = 11.84VQIRR729 pKa = 11.84KK730 pKa = 7.91MALVKK735 pKa = 10.76
Molecular weight: 79.84 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B0VGK6|B0VGK6_CLOAI Transcription-repair-coupling factor OS=Cloacimonas acidaminovorans (strain Evry) OX=459349 GN=mfd PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.3QPSNRR10 pKa = 11.84SRR12 pKa = 11.84KK13 pKa = 7.07NTHH16 pKa = 5.33GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.46NGRR28 pKa = 11.84KK29 pKa = 8.96VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.07GRR39 pKa = 11.84KK40 pKa = 6.31TLTVV44 pKa = 3.22
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.3QPSNRR10 pKa = 11.84SRR12 pKa = 11.84KK13 pKa = 7.07NTHH16 pKa = 5.33GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.46NGRR28 pKa = 11.84KK29 pKa = 8.96VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.07GRR39 pKa = 11.84KK40 pKa = 6.31TLTVV44 pKa = 3.22
Molecular weight: 5.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
649216 |
24 |
6457 |
358.1 |
40.48 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.513 ± 0.062 | 1.13 ± 0.022 |
5.034 ± 0.041 | 6.593 ± 0.067 |
4.552 ± 0.041 | 6.145 ± 0.055 |
1.677 ± 0.021 | 8.435 ± 0.049 |
6.825 ± 0.087 | 10.042 ± 0.086 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.247 ± 0.028 | 5.837 ± 0.064 |
4.268 ± 0.039 | 3.7 ± 0.035 |
3.884 ± 0.042 | 6.273 ± 0.054 |
5.539 ± 0.102 | 5.61 ± 0.055 |
1.157 ± 0.027 | 4.538 ± 0.049 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
