
Sclerotinia nivalis victorivirus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Totiviridae; Victorivirus; unclassified Victorivirus
Average proteome isoelectric point is 6.97
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A192ABA0|A0A192ABA0_9VIRU Coat protein OS=Sclerotinia nivalis victorivirus 1 OX=1859161 PE=4 SV=1
MM1 pKa = 6.39TTVIRR6 pKa = 11.84NSFLASVLASPRR18 pKa = 11.84GGYY21 pKa = 9.18IEE23 pKa = 5.12NDD25 pKa = 2.75NTYY28 pKa = 10.67RR29 pKa = 11.84RR30 pKa = 11.84YY31 pKa = 10.13KK32 pKa = 10.61SNVRR36 pKa = 11.84TSSTIGGNEE45 pKa = 4.15DD46 pKa = 3.19SRR48 pKa = 11.84LVQILYY54 pKa = 10.31EE55 pKa = 4.16VGRR58 pKa = 11.84SKK60 pKa = 11.0NSKK63 pKa = 9.32GRR65 pKa = 11.84ALARR69 pKa = 11.84PGEE72 pKa = 4.51DD73 pKa = 4.0DD74 pKa = 3.94PLIEE78 pKa = 5.42AGYY81 pKa = 7.58PTANALAEE89 pKa = 4.31DD90 pKa = 4.43FVGLSKK96 pKa = 11.09KK97 pKa = 8.47FTNFSATFEE106 pKa = 4.39FSSHH110 pKa = 4.97AAIVEE115 pKa = 3.99RR116 pKa = 11.84LARR119 pKa = 11.84GLAASSIFDD128 pKa = 4.45DD129 pKa = 4.23VDD131 pKa = 4.17SSDD134 pKa = 3.61LRR136 pKa = 11.84AGRR139 pKa = 11.84TLVINALGTYY149 pKa = 9.44DD150 pKa = 4.16GPVNSLTSSVFIPRR164 pKa = 11.84LVNSAITGDD173 pKa = 3.97VFSVLANAAAGEE185 pKa = 4.22GAEE188 pKa = 4.13IATDD192 pKa = 4.16IIEE195 pKa = 4.7LDD197 pKa = 3.3ALTRR201 pKa = 11.84QPIVPEE207 pKa = 4.2VEE209 pKa = 3.75ATALSRR215 pKa = 11.84AIVDD219 pKa = 3.5ALRR222 pKa = 11.84ILGANMIACDD232 pKa = 3.89QGPLFALALTRR243 pKa = 11.84GLHH246 pKa = 5.82KK247 pKa = 10.54VLTVVGHH254 pKa = 5.34TDD256 pKa = 2.95EE257 pKa = 5.08GAVSRR262 pKa = 11.84DD263 pKa = 3.37LLRR266 pKa = 11.84CGSFSVPFGDD276 pKa = 2.61IHH278 pKa = 7.98YY279 pKa = 10.37GLEE282 pKa = 4.29PYY284 pKa = 10.67AGIPALSTNSSYY296 pKa = 11.14GRR298 pKa = 11.84AAYY301 pKa = 10.14VDD303 pKa = 3.91SLALSTAALVAHH315 pKa = 7.18CDD317 pKa = 3.1PGQIYY322 pKa = 10.24DD323 pKa = 4.17GRR325 pKa = 11.84WLPTFFSGSSSTDD338 pKa = 2.77TDD340 pKa = 3.31GRR342 pKa = 11.84PGDD345 pKa = 4.06SLDD348 pKa = 3.64PTDD351 pKa = 5.98AMTLRR356 pKa = 11.84NRR358 pKa = 11.84AQLLSDD364 pKa = 4.16LPKK367 pKa = 10.6FSLSYY372 pKa = 10.61IPALAKK378 pKa = 10.65LFAAEE383 pKa = 4.38GNPAIAIAAFNGASSQLSRR402 pKa = 11.84TTRR405 pKa = 11.84HH406 pKa = 5.84LKK408 pKa = 9.56FASVSPWFWIEE419 pKa = 3.71PTSLIPHH426 pKa = 6.98DD427 pKa = 4.16FVGSVAEE434 pKa = 4.21HH435 pKa = 6.68EE436 pKa = 4.87GFASYY441 pKa = 10.7GGRR444 pKa = 11.84DD445 pKa = 3.52STRR448 pKa = 11.84SRR450 pKa = 11.84HH451 pKa = 4.49AWEE454 pKa = 5.9DD455 pKa = 3.34IQASGVNDD463 pKa = 3.74TAFSAYY469 pKa = 9.58HH470 pKa = 5.77ALFRR474 pKa = 11.84GARR477 pKa = 11.84ACWFFLHH484 pKa = 6.71WLGNPANGLGSISVRR499 pKa = 11.84QLDD502 pKa = 4.15PNAIIHH508 pKa = 6.61PGPCTAHH515 pKa = 4.78QQVRR519 pKa = 11.84DD520 pKa = 3.62RR521 pKa = 11.84VEE523 pKa = 5.12ASLPWTDD530 pKa = 3.99YY531 pKa = 11.03LWTRR535 pKa = 11.84GQSPFPAPGEE545 pKa = 3.88MLNISGTVGFFVKK558 pKa = 10.2HH559 pKa = 4.56YY560 pKa = 9.25TFDD563 pKa = 4.77DD564 pKa = 3.78EE565 pKa = 4.83GLPTVEE571 pKa = 4.98HH572 pKa = 6.43VPASHH577 pKa = 6.75EE578 pKa = 4.13FLDD581 pKa = 4.13TTVTLHH587 pKa = 5.32VGRR590 pKa = 11.84PIGLSIGASNTPASDD605 pKa = 2.87ARR607 pKa = 11.84RR608 pKa = 11.84ARR610 pKa = 11.84TRR612 pKa = 11.84ATRR615 pKa = 11.84EE616 pKa = 3.46LAAASARR623 pKa = 11.84AAVFGRR629 pKa = 11.84PDD631 pKa = 3.39VSEE634 pKa = 4.32MPTLTTAPVLRR645 pKa = 11.84SRR647 pKa = 11.84NPTLPTHH654 pKa = 6.11ITPNDD659 pKa = 3.78TPGGTDD665 pKa = 2.98VTHH668 pKa = 7.51RR669 pKa = 11.84SRR671 pKa = 11.84HH672 pKa = 4.79QGVGGADD679 pKa = 3.05ASDD682 pKa = 3.77RR683 pKa = 11.84LPAGEE688 pKa = 4.26PRR690 pKa = 11.84IPVPQHH696 pKa = 4.65QPVRR700 pKa = 11.84YY701 pKa = 7.85PQVPRR706 pKa = 11.84PAGNQPGGGGPGIPPAPPGPGGPNGDD732 pKa = 5.15DD733 pKa = 5.05DD734 pKa = 6.67DD735 pKa = 5.44NDD737 pKa = 3.58PPAPVNPPEE746 pKa = 4.98GGPPTPPAGAAPPNGPAPII765 pKa = 4.49
MM1 pKa = 6.39TTVIRR6 pKa = 11.84NSFLASVLASPRR18 pKa = 11.84GGYY21 pKa = 9.18IEE23 pKa = 5.12NDD25 pKa = 2.75NTYY28 pKa = 10.67RR29 pKa = 11.84RR30 pKa = 11.84YY31 pKa = 10.13KK32 pKa = 10.61SNVRR36 pKa = 11.84TSSTIGGNEE45 pKa = 4.15DD46 pKa = 3.19SRR48 pKa = 11.84LVQILYY54 pKa = 10.31EE55 pKa = 4.16VGRR58 pKa = 11.84SKK60 pKa = 11.0NSKK63 pKa = 9.32GRR65 pKa = 11.84ALARR69 pKa = 11.84PGEE72 pKa = 4.51DD73 pKa = 4.0DD74 pKa = 3.94PLIEE78 pKa = 5.42AGYY81 pKa = 7.58PTANALAEE89 pKa = 4.31DD90 pKa = 4.43FVGLSKK96 pKa = 11.09KK97 pKa = 8.47FTNFSATFEE106 pKa = 4.39FSSHH110 pKa = 4.97AAIVEE115 pKa = 3.99RR116 pKa = 11.84LARR119 pKa = 11.84GLAASSIFDD128 pKa = 4.45DD129 pKa = 4.23VDD131 pKa = 4.17SSDD134 pKa = 3.61LRR136 pKa = 11.84AGRR139 pKa = 11.84TLVINALGTYY149 pKa = 9.44DD150 pKa = 4.16GPVNSLTSSVFIPRR164 pKa = 11.84LVNSAITGDD173 pKa = 3.97VFSVLANAAAGEE185 pKa = 4.22GAEE188 pKa = 4.13IATDD192 pKa = 4.16IIEE195 pKa = 4.7LDD197 pKa = 3.3ALTRR201 pKa = 11.84QPIVPEE207 pKa = 4.2VEE209 pKa = 3.75ATALSRR215 pKa = 11.84AIVDD219 pKa = 3.5ALRR222 pKa = 11.84ILGANMIACDD232 pKa = 3.89QGPLFALALTRR243 pKa = 11.84GLHH246 pKa = 5.82KK247 pKa = 10.54VLTVVGHH254 pKa = 5.34TDD256 pKa = 2.95EE257 pKa = 5.08GAVSRR262 pKa = 11.84DD263 pKa = 3.37LLRR266 pKa = 11.84CGSFSVPFGDD276 pKa = 2.61IHH278 pKa = 7.98YY279 pKa = 10.37GLEE282 pKa = 4.29PYY284 pKa = 10.67AGIPALSTNSSYY296 pKa = 11.14GRR298 pKa = 11.84AAYY301 pKa = 10.14VDD303 pKa = 3.91SLALSTAALVAHH315 pKa = 7.18CDD317 pKa = 3.1PGQIYY322 pKa = 10.24DD323 pKa = 4.17GRR325 pKa = 11.84WLPTFFSGSSSTDD338 pKa = 2.77TDD340 pKa = 3.31GRR342 pKa = 11.84PGDD345 pKa = 4.06SLDD348 pKa = 3.64PTDD351 pKa = 5.98AMTLRR356 pKa = 11.84NRR358 pKa = 11.84AQLLSDD364 pKa = 4.16LPKK367 pKa = 10.6FSLSYY372 pKa = 10.61IPALAKK378 pKa = 10.65LFAAEE383 pKa = 4.38GNPAIAIAAFNGASSQLSRR402 pKa = 11.84TTRR405 pKa = 11.84HH406 pKa = 5.84LKK408 pKa = 9.56FASVSPWFWIEE419 pKa = 3.71PTSLIPHH426 pKa = 6.98DD427 pKa = 4.16FVGSVAEE434 pKa = 4.21HH435 pKa = 6.68EE436 pKa = 4.87GFASYY441 pKa = 10.7GGRR444 pKa = 11.84DD445 pKa = 3.52STRR448 pKa = 11.84SRR450 pKa = 11.84HH451 pKa = 4.49AWEE454 pKa = 5.9DD455 pKa = 3.34IQASGVNDD463 pKa = 3.74TAFSAYY469 pKa = 9.58HH470 pKa = 5.77ALFRR474 pKa = 11.84GARR477 pKa = 11.84ACWFFLHH484 pKa = 6.71WLGNPANGLGSISVRR499 pKa = 11.84QLDD502 pKa = 4.15PNAIIHH508 pKa = 6.61PGPCTAHH515 pKa = 4.78QQVRR519 pKa = 11.84DD520 pKa = 3.62RR521 pKa = 11.84VEE523 pKa = 5.12ASLPWTDD530 pKa = 3.99YY531 pKa = 11.03LWTRR535 pKa = 11.84GQSPFPAPGEE545 pKa = 3.88MLNISGTVGFFVKK558 pKa = 10.2HH559 pKa = 4.56YY560 pKa = 9.25TFDD563 pKa = 4.77DD564 pKa = 3.78EE565 pKa = 4.83GLPTVEE571 pKa = 4.98HH572 pKa = 6.43VPASHH577 pKa = 6.75EE578 pKa = 4.13FLDD581 pKa = 4.13TTVTLHH587 pKa = 5.32VGRR590 pKa = 11.84PIGLSIGASNTPASDD605 pKa = 2.87ARR607 pKa = 11.84RR608 pKa = 11.84ARR610 pKa = 11.84TRR612 pKa = 11.84ATRR615 pKa = 11.84EE616 pKa = 3.46LAAASARR623 pKa = 11.84AAVFGRR629 pKa = 11.84PDD631 pKa = 3.39VSEE634 pKa = 4.32MPTLTTAPVLRR645 pKa = 11.84SRR647 pKa = 11.84NPTLPTHH654 pKa = 6.11ITPNDD659 pKa = 3.78TPGGTDD665 pKa = 2.98VTHH668 pKa = 7.51RR669 pKa = 11.84SRR671 pKa = 11.84HH672 pKa = 4.79QGVGGADD679 pKa = 3.05ASDD682 pKa = 3.77RR683 pKa = 11.84LPAGEE688 pKa = 4.26PRR690 pKa = 11.84IPVPQHH696 pKa = 4.65QPVRR700 pKa = 11.84YY701 pKa = 7.85PQVPRR706 pKa = 11.84PAGNQPGGGGPGIPPAPPGPGGPNGDD732 pKa = 5.15DD733 pKa = 5.05DD734 pKa = 6.67DD735 pKa = 5.44NDD737 pKa = 3.58PPAPVNPPEE746 pKa = 4.98GGPPTPPAGAAPPNGPAPII765 pKa = 4.49
Molecular weight: 80.91 kDa
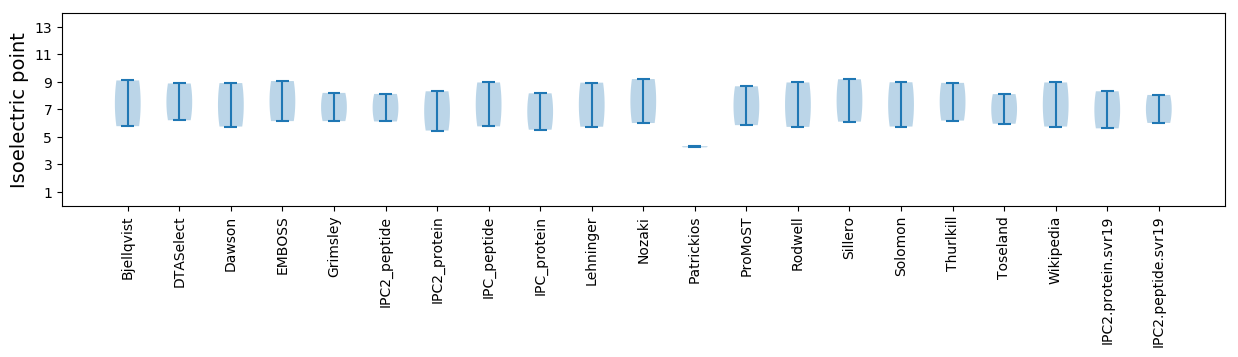
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A192ABA0|A0A192ABA0_9VIRU Coat protein OS=Sclerotinia nivalis victorivirus 1 OX=1859161 PE=4 SV=1
MM1 pKa = 7.15SAFVEE6 pKa = 4.35RR7 pKa = 11.84ASAYY11 pKa = 10.24GVVGDD16 pKa = 4.29YY17 pKa = 11.1LLSLRR22 pKa = 11.84SYY24 pKa = 10.91FPDD27 pKa = 3.16SFKK30 pKa = 11.13GYY32 pKa = 10.17GQTTFVNDD40 pKa = 3.72LVRR43 pKa = 11.84LQSSTPTLKK52 pKa = 9.83TKK54 pKa = 10.43HH55 pKa = 6.39ALLPAAVSLLALPFPLQVPLTLSDD79 pKa = 3.39ILVLAKK85 pKa = 10.12RR86 pKa = 11.84ASGVPFDD93 pKa = 4.48YY94 pKa = 10.81VHH96 pKa = 6.46SARR99 pKa = 11.84WSDD102 pKa = 3.61SPLISSEE109 pKa = 3.76YY110 pKa = 9.68RR111 pKa = 11.84GRR113 pKa = 11.84DD114 pKa = 3.53YY115 pKa = 11.03MVLCTNALFLRR126 pKa = 11.84NCFPVKK132 pKa = 9.93KK133 pKa = 9.65HH134 pKa = 5.64ASAYY138 pKa = 9.65SKK140 pKa = 11.42VNVSLPDD147 pKa = 3.74LAVSLCKK154 pKa = 10.32LGHH157 pKa = 5.15TRR159 pKa = 11.84SLIGMIFRR167 pKa = 11.84CAGLLHH173 pKa = 7.17EE174 pKa = 5.06DD175 pKa = 3.36AFMNALIYY183 pKa = 10.36GYY185 pKa = 10.45ALSYY189 pKa = 10.99HH190 pKa = 6.63FGASGWDD197 pKa = 3.52IACHH201 pKa = 6.59FILNPKK207 pKa = 8.26IAKK210 pKa = 9.28NVSVSLKK217 pKa = 10.77ALGANTTRR225 pKa = 11.84EE226 pKa = 3.87GALLVEE232 pKa = 5.02ADD234 pKa = 3.82TLQGRR239 pKa = 11.84LAAPVDD245 pKa = 3.22IDD247 pKa = 3.21AQARR251 pKa = 11.84YY252 pKa = 9.47RR253 pKa = 11.84CDD255 pKa = 3.09PVAVAKK261 pKa = 10.55SVIPYY266 pKa = 9.63SDD268 pKa = 3.81EE269 pKa = 3.79LRR271 pKa = 11.84THH273 pKa = 6.16IRR275 pKa = 11.84AILLDD280 pKa = 3.52EE281 pKa = 4.41TRR283 pKa = 11.84DD284 pKa = 3.91RR285 pKa = 11.84EE286 pKa = 4.41CVLPNLHH293 pKa = 6.38SWWSARR299 pKa = 11.84WLWCVNGSQTTKK311 pKa = 10.36SSKK314 pKa = 11.29DD315 pKa = 3.11MGLDD319 pKa = 3.4PKK321 pKa = 11.11ANRR324 pKa = 11.84ATHH327 pKa = 6.41DD328 pKa = 3.17QDD330 pKa = 3.5FRR332 pKa = 11.84RR333 pKa = 11.84MASEE337 pKa = 3.9ALKK340 pKa = 10.74SEE342 pKa = 4.87PLTTWDD348 pKa = 3.29GTTSVSKK355 pKa = 10.59SIKK358 pKa = 10.22LEE360 pKa = 4.04NGKK363 pKa = 9.26EE364 pKa = 3.79RR365 pKa = 11.84AIFACDD371 pKa = 3.22TNSYY375 pKa = 10.54FAFSWLLSSAEE386 pKa = 4.08KK387 pKa = 9.61VWRR390 pKa = 11.84GKK392 pKa = 10.39RR393 pKa = 11.84VILDD397 pKa = 3.66PGVGGHH403 pKa = 7.04LGIAKK408 pKa = 9.74RR409 pKa = 11.84VQGAQRR415 pKa = 11.84GGGVNLMLDD424 pKa = 3.48YY425 pKa = 11.52DD426 pKa = 4.57DD427 pKa = 6.33FNSQHH432 pKa = 6.18ATPVMQCVIDD442 pKa = 3.92EE443 pKa = 4.39LCNIFSCPDD452 pKa = 3.05WYY454 pKa = 10.95RR455 pKa = 11.84EE456 pKa = 3.89LLINSLDD463 pKa = 3.52KK464 pKa = 11.0MFINIDD470 pKa = 3.45GAPQRR475 pKa = 11.84VVGTLMSGHH484 pKa = 7.12RR485 pKa = 11.84GTTFFNSVLNAAYY498 pKa = 9.5IRR500 pKa = 11.84LAVGASAFDD509 pKa = 3.79SLISLHH515 pKa = 6.98AGDD518 pKa = 5.56DD519 pKa = 3.74VYY521 pKa = 11.1IRR523 pKa = 11.84CNTLSDD529 pKa = 4.29CDD531 pKa = 4.37SILNNTRR538 pKa = 11.84RR539 pKa = 11.84VGCRR543 pKa = 11.84MNPSKK548 pKa = 10.84QSIGFVGAEE557 pKa = 4.07FLRR560 pKa = 11.84VGIGTNAAYY569 pKa = 9.93GYY571 pKa = 9.27LARR574 pKa = 11.84GIAGFVSGNWVTQDD588 pKa = 3.17ILAPDD593 pKa = 4.3DD594 pKa = 4.43ALVSSISACRR604 pKa = 11.84TLINRR609 pKa = 11.84SGCASLALLLSRR621 pKa = 11.84SLRR624 pKa = 11.84YY625 pKa = 8.31TSGFSHH631 pKa = 6.75RR632 pKa = 11.84VLRR635 pKa = 11.84SLFSGEE641 pKa = 4.13SALEE645 pKa = 3.82GSPVYY650 pKa = 10.73NVDD653 pKa = 3.38YY654 pKa = 10.79QIRR657 pKa = 11.84NYY659 pKa = 10.64KK660 pKa = 8.16MNRR663 pKa = 11.84VTPDD667 pKa = 3.88SIPVPSNWARR677 pKa = 11.84HH678 pKa = 4.78ASIDD682 pKa = 3.78YY683 pKa = 7.64LTSHH687 pKa = 7.05LSEE690 pKa = 5.19IEE692 pKa = 3.92VTAINMVGVDD702 pKa = 4.06ALNLLVTSSYY712 pKa = 11.7SKK714 pKa = 10.65GLNMKK719 pKa = 9.46YY720 pKa = 9.67RR721 pKa = 11.84AALTRR726 pKa = 11.84PTISRR731 pKa = 11.84MTVRR735 pKa = 11.84LARR738 pKa = 11.84GFVSAATLLDD748 pKa = 3.17KK749 pKa = 10.83RR750 pKa = 11.84LEE752 pKa = 4.38RR753 pKa = 11.84GCLSQYY759 pKa = 9.98PVVRR763 pKa = 11.84LFEE766 pKa = 4.15NRR768 pKa = 11.84LDD770 pKa = 3.94HH771 pKa = 7.2NSLRR775 pKa = 11.84EE776 pKa = 4.23LITLAGGNPAARR788 pKa = 11.84DD789 pKa = 3.55LRR791 pKa = 11.84EE792 pKa = 3.83EE793 pKa = 4.1AFGRR797 pKa = 11.84TSLSKK802 pKa = 10.99NIIGYY807 pKa = 9.82LPFGDD812 pKa = 3.9AASLSKK818 pKa = 10.19RR819 pKa = 11.84TTAGNIFTLYY829 pKa = 10.56NVYY832 pKa = 8.82TT833 pKa = 3.96
MM1 pKa = 7.15SAFVEE6 pKa = 4.35RR7 pKa = 11.84ASAYY11 pKa = 10.24GVVGDD16 pKa = 4.29YY17 pKa = 11.1LLSLRR22 pKa = 11.84SYY24 pKa = 10.91FPDD27 pKa = 3.16SFKK30 pKa = 11.13GYY32 pKa = 10.17GQTTFVNDD40 pKa = 3.72LVRR43 pKa = 11.84LQSSTPTLKK52 pKa = 9.83TKK54 pKa = 10.43HH55 pKa = 6.39ALLPAAVSLLALPFPLQVPLTLSDD79 pKa = 3.39ILVLAKK85 pKa = 10.12RR86 pKa = 11.84ASGVPFDD93 pKa = 4.48YY94 pKa = 10.81VHH96 pKa = 6.46SARR99 pKa = 11.84WSDD102 pKa = 3.61SPLISSEE109 pKa = 3.76YY110 pKa = 9.68RR111 pKa = 11.84GRR113 pKa = 11.84DD114 pKa = 3.53YY115 pKa = 11.03MVLCTNALFLRR126 pKa = 11.84NCFPVKK132 pKa = 9.93KK133 pKa = 9.65HH134 pKa = 5.64ASAYY138 pKa = 9.65SKK140 pKa = 11.42VNVSLPDD147 pKa = 3.74LAVSLCKK154 pKa = 10.32LGHH157 pKa = 5.15TRR159 pKa = 11.84SLIGMIFRR167 pKa = 11.84CAGLLHH173 pKa = 7.17EE174 pKa = 5.06DD175 pKa = 3.36AFMNALIYY183 pKa = 10.36GYY185 pKa = 10.45ALSYY189 pKa = 10.99HH190 pKa = 6.63FGASGWDD197 pKa = 3.52IACHH201 pKa = 6.59FILNPKK207 pKa = 8.26IAKK210 pKa = 9.28NVSVSLKK217 pKa = 10.77ALGANTTRR225 pKa = 11.84EE226 pKa = 3.87GALLVEE232 pKa = 5.02ADD234 pKa = 3.82TLQGRR239 pKa = 11.84LAAPVDD245 pKa = 3.22IDD247 pKa = 3.21AQARR251 pKa = 11.84YY252 pKa = 9.47RR253 pKa = 11.84CDD255 pKa = 3.09PVAVAKK261 pKa = 10.55SVIPYY266 pKa = 9.63SDD268 pKa = 3.81EE269 pKa = 3.79LRR271 pKa = 11.84THH273 pKa = 6.16IRR275 pKa = 11.84AILLDD280 pKa = 3.52EE281 pKa = 4.41TRR283 pKa = 11.84DD284 pKa = 3.91RR285 pKa = 11.84EE286 pKa = 4.41CVLPNLHH293 pKa = 6.38SWWSARR299 pKa = 11.84WLWCVNGSQTTKK311 pKa = 10.36SSKK314 pKa = 11.29DD315 pKa = 3.11MGLDD319 pKa = 3.4PKK321 pKa = 11.11ANRR324 pKa = 11.84ATHH327 pKa = 6.41DD328 pKa = 3.17QDD330 pKa = 3.5FRR332 pKa = 11.84RR333 pKa = 11.84MASEE337 pKa = 3.9ALKK340 pKa = 10.74SEE342 pKa = 4.87PLTTWDD348 pKa = 3.29GTTSVSKK355 pKa = 10.59SIKK358 pKa = 10.22LEE360 pKa = 4.04NGKK363 pKa = 9.26EE364 pKa = 3.79RR365 pKa = 11.84AIFACDD371 pKa = 3.22TNSYY375 pKa = 10.54FAFSWLLSSAEE386 pKa = 4.08KK387 pKa = 9.61VWRR390 pKa = 11.84GKK392 pKa = 10.39RR393 pKa = 11.84VILDD397 pKa = 3.66PGVGGHH403 pKa = 7.04LGIAKK408 pKa = 9.74RR409 pKa = 11.84VQGAQRR415 pKa = 11.84GGGVNLMLDD424 pKa = 3.48YY425 pKa = 11.52DD426 pKa = 4.57DD427 pKa = 6.33FNSQHH432 pKa = 6.18ATPVMQCVIDD442 pKa = 3.92EE443 pKa = 4.39LCNIFSCPDD452 pKa = 3.05WYY454 pKa = 10.95RR455 pKa = 11.84EE456 pKa = 3.89LLINSLDD463 pKa = 3.52KK464 pKa = 11.0MFINIDD470 pKa = 3.45GAPQRR475 pKa = 11.84VVGTLMSGHH484 pKa = 7.12RR485 pKa = 11.84GTTFFNSVLNAAYY498 pKa = 9.5IRR500 pKa = 11.84LAVGASAFDD509 pKa = 3.79SLISLHH515 pKa = 6.98AGDD518 pKa = 5.56DD519 pKa = 3.74VYY521 pKa = 11.1IRR523 pKa = 11.84CNTLSDD529 pKa = 4.29CDD531 pKa = 4.37SILNNTRR538 pKa = 11.84RR539 pKa = 11.84VGCRR543 pKa = 11.84MNPSKK548 pKa = 10.84QSIGFVGAEE557 pKa = 4.07FLRR560 pKa = 11.84VGIGTNAAYY569 pKa = 9.93GYY571 pKa = 9.27LARR574 pKa = 11.84GIAGFVSGNWVTQDD588 pKa = 3.17ILAPDD593 pKa = 4.3DD594 pKa = 4.43ALVSSISACRR604 pKa = 11.84TLINRR609 pKa = 11.84SGCASLALLLSRR621 pKa = 11.84SLRR624 pKa = 11.84YY625 pKa = 8.31TSGFSHH631 pKa = 6.75RR632 pKa = 11.84VLRR635 pKa = 11.84SLFSGEE641 pKa = 4.13SALEE645 pKa = 3.82GSPVYY650 pKa = 10.73NVDD653 pKa = 3.38YY654 pKa = 10.79QIRR657 pKa = 11.84NYY659 pKa = 10.64KK660 pKa = 8.16MNRR663 pKa = 11.84VTPDD667 pKa = 3.88SIPVPSNWARR677 pKa = 11.84HH678 pKa = 4.78ASIDD682 pKa = 3.78YY683 pKa = 7.64LTSHH687 pKa = 7.05LSEE690 pKa = 5.19IEE692 pKa = 3.92VTAINMVGVDD702 pKa = 4.06ALNLLVTSSYY712 pKa = 11.7SKK714 pKa = 10.65GLNMKK719 pKa = 9.46YY720 pKa = 9.67RR721 pKa = 11.84AALTRR726 pKa = 11.84PTISRR731 pKa = 11.84MTVRR735 pKa = 11.84LARR738 pKa = 11.84GFVSAATLLDD748 pKa = 3.17KK749 pKa = 10.83RR750 pKa = 11.84LEE752 pKa = 4.38RR753 pKa = 11.84GCLSQYY759 pKa = 9.98PVVRR763 pKa = 11.84LFEE766 pKa = 4.15NRR768 pKa = 11.84LDD770 pKa = 3.94HH771 pKa = 7.2NSLRR775 pKa = 11.84EE776 pKa = 4.23LITLAGGNPAARR788 pKa = 11.84DD789 pKa = 3.55LRR791 pKa = 11.84EE792 pKa = 3.83EE793 pKa = 4.1AFGRR797 pKa = 11.84TSLSKK802 pKa = 10.99NIIGYY807 pKa = 9.82LPFGDD812 pKa = 3.9AASLSKK818 pKa = 10.19RR819 pKa = 11.84TTAGNIFTLYY829 pKa = 10.56NVYY832 pKa = 8.82TT833 pKa = 3.96
Molecular weight: 91.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1598 |
765 |
833 |
799.0 |
86.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.513 ± 0.856 | 1.439 ± 0.537 |
5.945 ± 0.225 | 3.379 ± 0.282 |
3.942 ± 0.075 | 8.073 ± 0.916 |
2.441 ± 0.208 | 4.819 ± 0.077 |
2.503 ± 0.818 | 10.013 ± 1.305 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.252 ± 0.409 | 4.38 ± 0.314 |
6.32 ± 1.936 | 2.003 ± 0.061 |
7.009 ± 0.145 | 9.199 ± 0.48 |
6.07 ± 0.497 | 6.446 ± 0.296 |
1.252 ± 0.141 | 3.004 ± 0.535 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
