
Flaviflexus salsibiostraticola
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Actinomycetales; Actinomycetaceae; Flaviflexus
Average proteome isoelectric point is 5.81
Get precalculated fractions of proteins
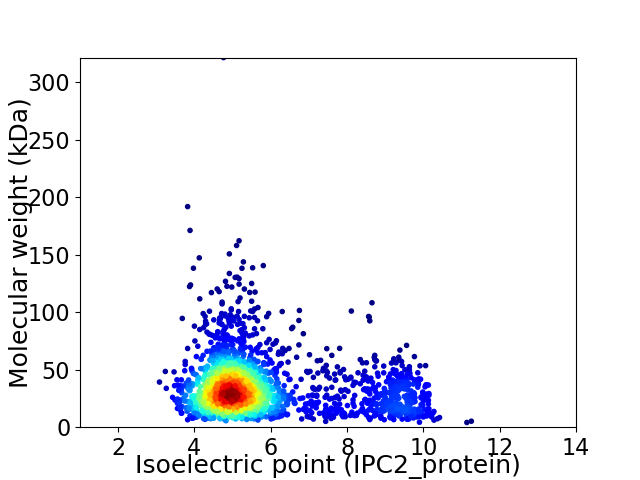
Virtual 2D-PAGE plot for 2276 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3Q8WUD4|A0A3Q8WUD4_9ACTO Uncharacterized protein OS=Flaviflexus salsibiostraticola OX=1282737 GN=EJO69_09615 PE=4 SV=1
MM1 pKa = 7.34KK2 pKa = 9.07RR3 pKa = 11.84TLAALASLALLLGACSSTAEE23 pKa = 4.19EE24 pKa = 4.41TASDD28 pKa = 3.94TADD31 pKa = 3.47SPVPTQPAPAGLEE44 pKa = 3.79EE45 pKa = 5.3FYY47 pKa = 11.1NQSLEE52 pKa = 3.9WGEE55 pKa = 4.59CMDD58 pKa = 3.86GTSAEE63 pKa = 4.16AEE65 pKa = 4.33CAKK68 pKa = 10.42VRR70 pKa = 11.84VPLDD74 pKa = 3.42YY75 pKa = 10.78EE76 pKa = 4.61DD77 pKa = 4.77PSEE80 pKa = 4.06ATIDD84 pKa = 3.5IAVLRR89 pKa = 11.84IPAGGEE95 pKa = 3.86AQGSLVVNPGGPGASGMDD113 pKa = 3.48MADD116 pKa = 2.9SAGAYY121 pKa = 9.34FSRR124 pKa = 11.84DD125 pKa = 3.11LVEE128 pKa = 5.04SYY130 pKa = 10.88DD131 pKa = 4.22IIGFDD136 pKa = 3.43PRR138 pKa = 11.84GVGEE142 pKa = 4.36SAPVDD147 pKa = 3.73CLPDD151 pKa = 3.69EE152 pKa = 4.46EE153 pKa = 6.24LGDD156 pKa = 4.24LLDD159 pKa = 3.93RR160 pKa = 11.84AYY162 pKa = 10.0PDD164 pKa = 3.63TPEE167 pKa = 5.7GEE169 pKa = 4.39AEE171 pKa = 4.02ALRR174 pKa = 11.84DD175 pKa = 3.58TKK177 pKa = 10.6TIISSCTKK185 pKa = 10.68ASGHH189 pKa = 5.72LLEE192 pKa = 5.38HH193 pKa = 7.05LSTINTARR201 pKa = 11.84DD202 pKa = 3.47MDD204 pKa = 3.85IMRR207 pKa = 11.84AALGEE212 pKa = 3.89PHH214 pKa = 7.28LDD216 pKa = 3.38YY217 pKa = 11.48LGFSYY222 pKa = 8.83GTHH225 pKa = 7.12LGGQYY230 pKa = 11.24ADD232 pKa = 4.62LFPEE236 pKa = 4.07NVGRR240 pKa = 11.84MVLDD244 pKa = 3.96GAIDD248 pKa = 3.71PAISSLDD255 pKa = 3.23SSYY258 pKa = 10.69FQAVGFEE265 pKa = 4.33KK266 pKa = 10.72ALEE269 pKa = 4.52AYY271 pKa = 10.44LEE273 pKa = 4.26YY274 pKa = 10.86CLEE277 pKa = 5.22DD278 pKa = 3.49EE279 pKa = 5.16DD280 pKa = 5.59CPLEE284 pKa = 4.75AGTVDD289 pKa = 3.37EE290 pKa = 5.75AKK292 pKa = 10.75AHH294 pKa = 5.45LQSEE298 pKa = 4.71IEE300 pKa = 4.3ASLEE304 pKa = 4.22SPIPTSIDD312 pKa = 3.3DD313 pKa = 3.77RR314 pKa = 11.84SVNPAIFYY322 pKa = 9.77TGIATTLYY330 pKa = 10.93SEE332 pKa = 4.48DD333 pKa = 2.97TWPFLTEE340 pKa = 3.97ALRR343 pKa = 11.84MAIEE347 pKa = 5.15DD348 pKa = 3.68GDD350 pKa = 4.14GTLLLTFNDD359 pKa = 3.78LMLGRR364 pKa = 11.84DD365 pKa = 3.57TFSGEE370 pKa = 3.94FMDD373 pKa = 5.24NSLEE377 pKa = 4.02ARR379 pKa = 11.84WAINCRR385 pKa = 11.84DD386 pKa = 3.99FPAEE390 pKa = 3.84TNAAAVEE397 pKa = 4.21ANDD400 pKa = 4.08ARR402 pKa = 11.84LNEE405 pKa = 4.55DD406 pKa = 3.89APTFAPFFEE415 pKa = 6.36GGDD418 pKa = 3.56IFCAGWPYY426 pKa = 10.1TADD429 pKa = 3.49EE430 pKa = 4.55VPGPFVAEE438 pKa = 4.32GSNPIVVIGTEE449 pKa = 4.37GDD451 pKa = 3.23PATPYY456 pKa = 9.75EE457 pKa = 4.2ASVNLAEE464 pKa = 4.05QLEE467 pKa = 4.18NGILVTWEE475 pKa = 4.54GEE477 pKa = 3.86GHH479 pKa = 5.39TAYY482 pKa = 8.92GTAGEE487 pKa = 5.09CIDD490 pKa = 4.83DD491 pKa = 3.86AVDD494 pKa = 3.78RR495 pKa = 11.84YY496 pKa = 10.53FIDD499 pKa = 4.71GEE501 pKa = 4.2LPEE504 pKa = 6.71DD505 pKa = 4.49GLRR508 pKa = 11.84CPAA511 pKa = 4.43
MM1 pKa = 7.34KK2 pKa = 9.07RR3 pKa = 11.84TLAALASLALLLGACSSTAEE23 pKa = 4.19EE24 pKa = 4.41TASDD28 pKa = 3.94TADD31 pKa = 3.47SPVPTQPAPAGLEE44 pKa = 3.79EE45 pKa = 5.3FYY47 pKa = 11.1NQSLEE52 pKa = 3.9WGEE55 pKa = 4.59CMDD58 pKa = 3.86GTSAEE63 pKa = 4.16AEE65 pKa = 4.33CAKK68 pKa = 10.42VRR70 pKa = 11.84VPLDD74 pKa = 3.42YY75 pKa = 10.78EE76 pKa = 4.61DD77 pKa = 4.77PSEE80 pKa = 4.06ATIDD84 pKa = 3.5IAVLRR89 pKa = 11.84IPAGGEE95 pKa = 3.86AQGSLVVNPGGPGASGMDD113 pKa = 3.48MADD116 pKa = 2.9SAGAYY121 pKa = 9.34FSRR124 pKa = 11.84DD125 pKa = 3.11LVEE128 pKa = 5.04SYY130 pKa = 10.88DD131 pKa = 4.22IIGFDD136 pKa = 3.43PRR138 pKa = 11.84GVGEE142 pKa = 4.36SAPVDD147 pKa = 3.73CLPDD151 pKa = 3.69EE152 pKa = 4.46EE153 pKa = 6.24LGDD156 pKa = 4.24LLDD159 pKa = 3.93RR160 pKa = 11.84AYY162 pKa = 10.0PDD164 pKa = 3.63TPEE167 pKa = 5.7GEE169 pKa = 4.39AEE171 pKa = 4.02ALRR174 pKa = 11.84DD175 pKa = 3.58TKK177 pKa = 10.6TIISSCTKK185 pKa = 10.68ASGHH189 pKa = 5.72LLEE192 pKa = 5.38HH193 pKa = 7.05LSTINTARR201 pKa = 11.84DD202 pKa = 3.47MDD204 pKa = 3.85IMRR207 pKa = 11.84AALGEE212 pKa = 3.89PHH214 pKa = 7.28LDD216 pKa = 3.38YY217 pKa = 11.48LGFSYY222 pKa = 8.83GTHH225 pKa = 7.12LGGQYY230 pKa = 11.24ADD232 pKa = 4.62LFPEE236 pKa = 4.07NVGRR240 pKa = 11.84MVLDD244 pKa = 3.96GAIDD248 pKa = 3.71PAISSLDD255 pKa = 3.23SSYY258 pKa = 10.69FQAVGFEE265 pKa = 4.33KK266 pKa = 10.72ALEE269 pKa = 4.52AYY271 pKa = 10.44LEE273 pKa = 4.26YY274 pKa = 10.86CLEE277 pKa = 5.22DD278 pKa = 3.49EE279 pKa = 5.16DD280 pKa = 5.59CPLEE284 pKa = 4.75AGTVDD289 pKa = 3.37EE290 pKa = 5.75AKK292 pKa = 10.75AHH294 pKa = 5.45LQSEE298 pKa = 4.71IEE300 pKa = 4.3ASLEE304 pKa = 4.22SPIPTSIDD312 pKa = 3.3DD313 pKa = 3.77RR314 pKa = 11.84SVNPAIFYY322 pKa = 9.77TGIATTLYY330 pKa = 10.93SEE332 pKa = 4.48DD333 pKa = 2.97TWPFLTEE340 pKa = 3.97ALRR343 pKa = 11.84MAIEE347 pKa = 5.15DD348 pKa = 3.68GDD350 pKa = 4.14GTLLLTFNDD359 pKa = 3.78LMLGRR364 pKa = 11.84DD365 pKa = 3.57TFSGEE370 pKa = 3.94FMDD373 pKa = 5.24NSLEE377 pKa = 4.02ARR379 pKa = 11.84WAINCRR385 pKa = 11.84DD386 pKa = 3.99FPAEE390 pKa = 3.84TNAAAVEE397 pKa = 4.21ANDD400 pKa = 4.08ARR402 pKa = 11.84LNEE405 pKa = 4.55DD406 pKa = 3.89APTFAPFFEE415 pKa = 6.36GGDD418 pKa = 3.56IFCAGWPYY426 pKa = 10.1TADD429 pKa = 3.49EE430 pKa = 4.55VPGPFVAEE438 pKa = 4.32GSNPIVVIGTEE449 pKa = 4.37GDD451 pKa = 3.23PATPYY456 pKa = 9.75EE457 pKa = 4.2ASVNLAEE464 pKa = 4.05QLEE467 pKa = 4.18NGILVTWEE475 pKa = 4.54GEE477 pKa = 3.86GHH479 pKa = 5.39TAYY482 pKa = 8.92GTAGEE487 pKa = 5.09CIDD490 pKa = 4.83DD491 pKa = 3.86AVDD494 pKa = 3.78RR495 pKa = 11.84YY496 pKa = 10.53FIDD499 pKa = 4.71GEE501 pKa = 4.2LPEE504 pKa = 6.71DD505 pKa = 4.49GLRR508 pKa = 11.84CPAA511 pKa = 4.43
Molecular weight: 54.49 kDa
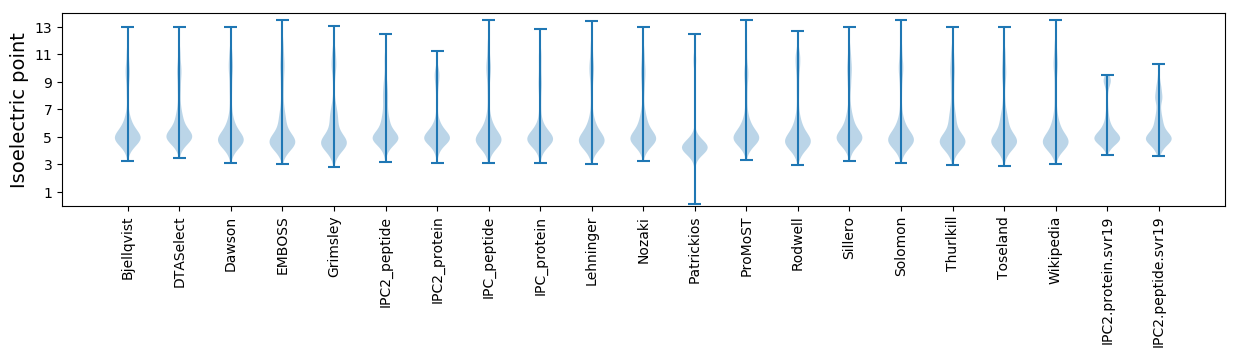
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3S8ZCP8|A0A3S8ZCP8_9ACTO ABC transporter ATP-binding protein OS=Flaviflexus salsibiostraticola OX=1282737 GN=EJO69_08750 PE=4 SV=1
MM1 pKa = 7.41VKK3 pKa = 9.03RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84KK12 pKa = 9.27RR13 pKa = 11.84SKK15 pKa = 9.5VHH17 pKa = 5.91GFRR20 pKa = 11.84KK21 pKa = 10.02RR22 pKa = 11.84MATRR26 pKa = 11.84AGRR29 pKa = 11.84AILAGRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.89GRR40 pKa = 11.84AKK42 pKa = 10.68LSAA45 pKa = 3.92
MM1 pKa = 7.41VKK3 pKa = 9.03RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84KK12 pKa = 9.27RR13 pKa = 11.84SKK15 pKa = 9.5VHH17 pKa = 5.91GFRR20 pKa = 11.84KK21 pKa = 10.02RR22 pKa = 11.84MATRR26 pKa = 11.84AGRR29 pKa = 11.84AILAGRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.89GRR40 pKa = 11.84AKK42 pKa = 10.68LSAA45 pKa = 3.92
Molecular weight: 5.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
752478 |
32 |
3034 |
330.6 |
35.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.796 ± 0.068 | 0.627 ± 0.015 |
6.321 ± 0.044 | 6.505 ± 0.047 |
3.014 ± 0.032 | 8.708 ± 0.041 |
2.105 ± 0.027 | 5.428 ± 0.037 |
2.401 ± 0.042 | 9.778 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.232 ± 0.022 | 2.217 ± 0.028 |
5.112 ± 0.036 | 2.781 ± 0.028 |
7.104 ± 0.052 | 6.003 ± 0.032 |
6.08 ± 0.039 | 8.158 ± 0.044 |
1.439 ± 0.02 | 2.189 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
