
Gordonia amarae NBRC 15530
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Corynebacteriales; Gordoniaceae; Gordonia; Gordonia amarae
Average proteome isoelectric point is 5.96
Get precalculated fractions of proteins
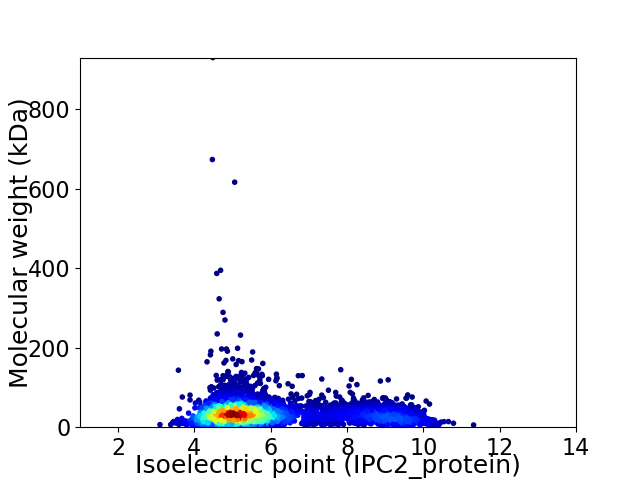
Virtual 2D-PAGE plot for 4691 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G7GNG8|G7GNG8_9ACTN DapB_N domain-containing protein OS=Gordonia amarae NBRC 15530 OX=1075090 GN=GOAMR_28_00300 PE=4 SV=1
MM1 pKa = 7.86RR2 pKa = 11.84ISHH5 pKa = 6.84KK6 pKa = 10.71NRR8 pKa = 11.84ARR10 pKa = 11.84LIAAAAGVSLFLNPGQAAADD30 pKa = 4.27DD31 pKa = 4.23DD32 pKa = 4.51TGSGTGSGASSHH44 pKa = 6.67SDD46 pKa = 2.75SSGGTNTSGGNSASGDD62 pKa = 3.92SQSTSSPGDD71 pKa = 3.54SNPAGNSGTTGGAGGSDD88 pKa = 3.48VTGGPDD94 pKa = 3.26STGGTDD100 pKa = 3.38NTASDD105 pKa = 4.65AGPSDD110 pKa = 3.94DD111 pKa = 4.26TDD113 pKa = 3.8SGSGSSPDD121 pKa = 3.39STGTSGNTTPADD133 pKa = 3.67TDD135 pKa = 3.76TTPSDD140 pKa = 3.6TTPADD145 pKa = 3.49AVPSGNEE152 pKa = 3.74TAGTTRR158 pKa = 11.84PPTEE162 pKa = 4.14TDD164 pKa = 2.88HH165 pKa = 6.85TEE167 pKa = 4.12PPAPADD173 pKa = 3.6TTVTPDD179 pKa = 3.29TTNTWTEE186 pKa = 3.87PTNTPTAPTTTGDD199 pKa = 3.67TTPTTSTAEE208 pKa = 4.15STSPDD213 pKa = 3.17ATGTADD219 pKa = 3.72TDD221 pKa = 3.74TTPPIGDD228 pKa = 3.9AEE230 pKa = 4.58TPGDD234 pKa = 3.5NSGPSEE240 pKa = 4.52PDD242 pKa = 2.96PTTLRR247 pKa = 11.84PLGFTAPAFAEE258 pKa = 4.4EE259 pKa = 4.82PEE261 pKa = 4.6TTGEE265 pKa = 4.24PEE267 pKa = 4.16TFSNAVFFAPPPATTPPAAPAPATPLTTLLTFFGIPPVGGPTAPTALTTPAPWTLLWWIQRR328 pKa = 11.84TQSLLNNQTPTATSVLDD345 pKa = 3.98PVTDD349 pKa = 4.76GYY351 pKa = 9.74TISLTTDD358 pKa = 3.55DD359 pKa = 5.28ADD361 pKa = 4.06GDD363 pKa = 4.28PVATTVYY370 pKa = 9.81HH371 pKa = 5.81QPTHH375 pKa = 5.07GTVVKK380 pKa = 10.78NSDD383 pKa = 3.18GAFTYY388 pKa = 10.38TRR390 pKa = 11.84DD391 pKa = 3.94QNSSAADD398 pKa = 3.2QFTIKK403 pKa = 10.62VSDD406 pKa = 3.95NGPHH410 pKa = 5.66LHH412 pKa = 6.76GLATVIGLFTGHH424 pKa = 7.39DD425 pKa = 3.51PHH427 pKa = 6.16STYY430 pKa = 9.79VTVNIPEE437 pKa = 4.29GTAAPANSAPVIDD450 pKa = 5.78AIDD453 pKa = 3.89YY454 pKa = 10.51SPQGSDD460 pKa = 3.33GVITGTTTATDD471 pKa = 3.52HH472 pKa = 7.59DD473 pKa = 5.34GDD475 pKa = 4.41TVTYY479 pKa = 9.19TYY481 pKa = 10.23TWDD484 pKa = 3.57TTKK487 pKa = 11.1GAIDD491 pKa = 4.18YY492 pKa = 7.47DD493 pKa = 3.96TGTGAFTFTPTYY505 pKa = 7.62TARR508 pKa = 11.84HH509 pKa = 5.59AAASDD514 pKa = 3.22NATPGDD520 pKa = 3.56KK521 pKa = 10.93ALTVTVNASDD531 pKa = 3.76GTLSDD536 pKa = 4.04TEE538 pKa = 4.61NFTLDD543 pKa = 2.87ILAANEE549 pKa = 4.32SVASNAPTSPLGTIDD564 pKa = 4.68RR565 pKa = 11.84DD566 pKa = 3.74MNAATYY572 pKa = 10.96GNTTGSLSGIATDD585 pKa = 5.06ADD587 pKa = 4.23NDD589 pKa = 3.99TLAYY593 pKa = 10.47SVTTQGANGTVTIVGDD609 pKa = 3.97SFTYY613 pKa = 10.36TPDD616 pKa = 3.18SANRR620 pKa = 11.84AFTDD624 pKa = 3.56TFTVTVDD631 pKa = 3.8DD632 pKa = 4.35GHH634 pKa = 7.14GSTTDD639 pKa = 3.07VTITVDD645 pKa = 3.74TVPMYY650 pKa = 10.28YY651 pKa = 10.19HH652 pKa = 7.25HH653 pKa = 7.79IDD655 pKa = 3.42PTNQFWDD662 pKa = 3.9PSDD665 pKa = 4.23AGFLTTIPAIVNPTGVVDD683 pKa = 3.78YY684 pKa = 11.04HH685 pKa = 8.31VDD687 pKa = 3.19EE688 pKa = 4.8TGNIIVNNTGSDD700 pKa = 4.25PIMLWSITNDD710 pKa = 2.64EE711 pKa = 4.21VTALNAGGPATFEE724 pKa = 4.1ALVIASGSSATIPPRR739 pKa = 11.84AGSTPFIWDD748 pKa = 3.61QIILLQAPRR757 pKa = 11.84GAEE760 pKa = 4.29PILLGQIVLNDD771 pKa = 3.7GTILSDD777 pKa = 4.31DD778 pKa = 5.72GIPQTPVEE786 pKa = 4.41VTVTPGSNGAVNGVVNGHH804 pKa = 5.45QAGTTYY810 pKa = 10.49SVQGGTPTGGGYY822 pKa = 7.59LTPDD826 pKa = 3.23GGTVVFDD833 pKa = 3.73NSSGTFVFTPEE844 pKa = 5.57GEE846 pKa = 4.13DD847 pKa = 3.5QYY849 pKa = 11.15WAGGGGPTTQAFVILADD866 pKa = 4.63DD867 pKa = 4.34GVNPPRR873 pKa = 11.84AIAVTVPIDD882 pKa = 4.46PIDD885 pKa = 3.79LVVIDD890 pKa = 4.05MTGLNSTDD898 pKa = 3.23SPRR901 pKa = 11.84YY902 pKa = 9.21SYY904 pKa = 11.19GVSDD908 pKa = 3.71TSGTPILTVVDD919 pKa = 3.59RR920 pKa = 11.84TTAQSHH926 pKa = 6.9AIALPSGHH934 pKa = 7.4DD935 pKa = 3.4NTSTVTFSADD945 pKa = 2.43GGYY948 pKa = 10.8AHH950 pKa = 7.2ITNSGGSTVSVIDD963 pKa = 3.55LTTNQVIGTYY973 pKa = 9.88IVTDD977 pKa = 3.48NGDD980 pKa = 3.74GTKK983 pKa = 10.5SLAWTGGTPSGFVMASFVDD1002 pKa = 3.51NGPITGADD1010 pKa = 3.52FSIGAEE1016 pKa = 3.99GDD1018 pKa = 3.28QAIIVATQTDD1028 pKa = 3.86GTYY1031 pKa = 9.61TVYY1034 pKa = 10.8LADD1037 pKa = 3.75SRR1039 pKa = 11.84TGDD1042 pKa = 3.28RR1043 pKa = 11.84TALSTANPQNGTGTPTVTFIGSTGIALLADD1073 pKa = 4.03GTTVTSISVTDD1084 pKa = 3.79FAVLQQTVVGGDD1096 pKa = 3.5LAFDD1100 pKa = 3.43WVVNPNSQFAYY1111 pKa = 10.15GVTADD1116 pKa = 3.71PTSPVTSVTVVEE1128 pKa = 4.66LTTGTITEE1136 pKa = 4.22IAVPPTSAYY1145 pKa = 10.39AIDD1148 pKa = 4.17ANGSALYY1155 pKa = 9.45TYY1157 pKa = 10.93VLDD1160 pKa = 4.33PFTFSTQVTRR1170 pKa = 11.84TPFGDD1175 pKa = 3.3AGNAISVTVPGAVAGNVPEE1194 pKa = 4.77LSWSSFDD1201 pKa = 4.84DD1202 pKa = 3.55AGNYY1206 pKa = 9.93YY1207 pKa = 9.25LTTVDD1212 pKa = 3.47VSVVAGVPTLGAVKK1226 pKa = 10.08VHH1228 pKa = 6.31VFDD1231 pKa = 4.24QAMNEE1236 pKa = 3.73IDD1238 pKa = 4.09TIVTDD1243 pKa = 3.86VPSGQVAAMTSVADD1257 pKa = 3.6GTVYY1261 pKa = 11.13LMVADD1266 pKa = 5.01PGSAQNTYY1274 pKa = 9.45TIHH1277 pKa = 6.86RR1278 pKa = 11.84VSADD1282 pKa = 3.07GSVDD1286 pKa = 3.64DD1287 pKa = 5.35LADD1290 pKa = 3.68GVGWVTADD1298 pKa = 3.43QQHH1301 pKa = 5.7MFVLRR1306 pKa = 11.84TNDD1309 pKa = 2.85GSGYY1313 pKa = 8.6NTFSVLDD1320 pKa = 4.15LTTDD1324 pKa = 3.5TQSAPISVSTSYY1336 pKa = 11.14DD1337 pKa = 3.13GEE1339 pKa = 3.96FRR1341 pKa = 11.84VFEE1344 pKa = 4.26GKK1346 pKa = 10.21YY1347 pKa = 10.18LAIAYY1352 pKa = 7.87HH1353 pKa = 6.56AGSNVYY1359 pKa = 8.75VHH1361 pKa = 6.89ILDD1364 pKa = 3.58VDD1366 pKa = 4.01GNATDD1371 pKa = 4.09FVIGADD1377 pKa = 3.65DD1378 pKa = 4.26FEE1380 pKa = 4.74GLIYY1384 pKa = 10.57LDD1386 pKa = 4.49DD1387 pKa = 3.95QFSVITSGGGYY1398 pKa = 7.32TQATLIRR1405 pKa = 11.84AA1406 pKa = 3.85
MM1 pKa = 7.86RR2 pKa = 11.84ISHH5 pKa = 6.84KK6 pKa = 10.71NRR8 pKa = 11.84ARR10 pKa = 11.84LIAAAAGVSLFLNPGQAAADD30 pKa = 4.27DD31 pKa = 4.23DD32 pKa = 4.51TGSGTGSGASSHH44 pKa = 6.67SDD46 pKa = 2.75SSGGTNTSGGNSASGDD62 pKa = 3.92SQSTSSPGDD71 pKa = 3.54SNPAGNSGTTGGAGGSDD88 pKa = 3.48VTGGPDD94 pKa = 3.26STGGTDD100 pKa = 3.38NTASDD105 pKa = 4.65AGPSDD110 pKa = 3.94DD111 pKa = 4.26TDD113 pKa = 3.8SGSGSSPDD121 pKa = 3.39STGTSGNTTPADD133 pKa = 3.67TDD135 pKa = 3.76TTPSDD140 pKa = 3.6TTPADD145 pKa = 3.49AVPSGNEE152 pKa = 3.74TAGTTRR158 pKa = 11.84PPTEE162 pKa = 4.14TDD164 pKa = 2.88HH165 pKa = 6.85TEE167 pKa = 4.12PPAPADD173 pKa = 3.6TTVTPDD179 pKa = 3.29TTNTWTEE186 pKa = 3.87PTNTPTAPTTTGDD199 pKa = 3.67TTPTTSTAEE208 pKa = 4.15STSPDD213 pKa = 3.17ATGTADD219 pKa = 3.72TDD221 pKa = 3.74TTPPIGDD228 pKa = 3.9AEE230 pKa = 4.58TPGDD234 pKa = 3.5NSGPSEE240 pKa = 4.52PDD242 pKa = 2.96PTTLRR247 pKa = 11.84PLGFTAPAFAEE258 pKa = 4.4EE259 pKa = 4.82PEE261 pKa = 4.6TTGEE265 pKa = 4.24PEE267 pKa = 4.16TFSNAVFFAPPPATTPPAAPAPATPLTTLLTFFGIPPVGGPTAPTALTTPAPWTLLWWIQRR328 pKa = 11.84TQSLLNNQTPTATSVLDD345 pKa = 3.98PVTDD349 pKa = 4.76GYY351 pKa = 9.74TISLTTDD358 pKa = 3.55DD359 pKa = 5.28ADD361 pKa = 4.06GDD363 pKa = 4.28PVATTVYY370 pKa = 9.81HH371 pKa = 5.81QPTHH375 pKa = 5.07GTVVKK380 pKa = 10.78NSDD383 pKa = 3.18GAFTYY388 pKa = 10.38TRR390 pKa = 11.84DD391 pKa = 3.94QNSSAADD398 pKa = 3.2QFTIKK403 pKa = 10.62VSDD406 pKa = 3.95NGPHH410 pKa = 5.66LHH412 pKa = 6.76GLATVIGLFTGHH424 pKa = 7.39DD425 pKa = 3.51PHH427 pKa = 6.16STYY430 pKa = 9.79VTVNIPEE437 pKa = 4.29GTAAPANSAPVIDD450 pKa = 5.78AIDD453 pKa = 3.89YY454 pKa = 10.51SPQGSDD460 pKa = 3.33GVITGTTTATDD471 pKa = 3.52HH472 pKa = 7.59DD473 pKa = 5.34GDD475 pKa = 4.41TVTYY479 pKa = 9.19TYY481 pKa = 10.23TWDD484 pKa = 3.57TTKK487 pKa = 11.1GAIDD491 pKa = 4.18YY492 pKa = 7.47DD493 pKa = 3.96TGTGAFTFTPTYY505 pKa = 7.62TARR508 pKa = 11.84HH509 pKa = 5.59AAASDD514 pKa = 3.22NATPGDD520 pKa = 3.56KK521 pKa = 10.93ALTVTVNASDD531 pKa = 3.76GTLSDD536 pKa = 4.04TEE538 pKa = 4.61NFTLDD543 pKa = 2.87ILAANEE549 pKa = 4.32SVASNAPTSPLGTIDD564 pKa = 4.68RR565 pKa = 11.84DD566 pKa = 3.74MNAATYY572 pKa = 10.96GNTTGSLSGIATDD585 pKa = 5.06ADD587 pKa = 4.23NDD589 pKa = 3.99TLAYY593 pKa = 10.47SVTTQGANGTVTIVGDD609 pKa = 3.97SFTYY613 pKa = 10.36TPDD616 pKa = 3.18SANRR620 pKa = 11.84AFTDD624 pKa = 3.56TFTVTVDD631 pKa = 3.8DD632 pKa = 4.35GHH634 pKa = 7.14GSTTDD639 pKa = 3.07VTITVDD645 pKa = 3.74TVPMYY650 pKa = 10.28YY651 pKa = 10.19HH652 pKa = 7.25HH653 pKa = 7.79IDD655 pKa = 3.42PTNQFWDD662 pKa = 3.9PSDD665 pKa = 4.23AGFLTTIPAIVNPTGVVDD683 pKa = 3.78YY684 pKa = 11.04HH685 pKa = 8.31VDD687 pKa = 3.19EE688 pKa = 4.8TGNIIVNNTGSDD700 pKa = 4.25PIMLWSITNDD710 pKa = 2.64EE711 pKa = 4.21VTALNAGGPATFEE724 pKa = 4.1ALVIASGSSATIPPRR739 pKa = 11.84AGSTPFIWDD748 pKa = 3.61QIILLQAPRR757 pKa = 11.84GAEE760 pKa = 4.29PILLGQIVLNDD771 pKa = 3.7GTILSDD777 pKa = 4.31DD778 pKa = 5.72GIPQTPVEE786 pKa = 4.41VTVTPGSNGAVNGVVNGHH804 pKa = 5.45QAGTTYY810 pKa = 10.49SVQGGTPTGGGYY822 pKa = 7.59LTPDD826 pKa = 3.23GGTVVFDD833 pKa = 3.73NSSGTFVFTPEE844 pKa = 5.57GEE846 pKa = 4.13DD847 pKa = 3.5QYY849 pKa = 11.15WAGGGGPTTQAFVILADD866 pKa = 4.63DD867 pKa = 4.34GVNPPRR873 pKa = 11.84AIAVTVPIDD882 pKa = 4.46PIDD885 pKa = 3.79LVVIDD890 pKa = 4.05MTGLNSTDD898 pKa = 3.23SPRR901 pKa = 11.84YY902 pKa = 9.21SYY904 pKa = 11.19GVSDD908 pKa = 3.71TSGTPILTVVDD919 pKa = 3.59RR920 pKa = 11.84TTAQSHH926 pKa = 6.9AIALPSGHH934 pKa = 7.4DD935 pKa = 3.4NTSTVTFSADD945 pKa = 2.43GGYY948 pKa = 10.8AHH950 pKa = 7.2ITNSGGSTVSVIDD963 pKa = 3.55LTTNQVIGTYY973 pKa = 9.88IVTDD977 pKa = 3.48NGDD980 pKa = 3.74GTKK983 pKa = 10.5SLAWTGGTPSGFVMASFVDD1002 pKa = 3.51NGPITGADD1010 pKa = 3.52FSIGAEE1016 pKa = 3.99GDD1018 pKa = 3.28QAIIVATQTDD1028 pKa = 3.86GTYY1031 pKa = 9.61TVYY1034 pKa = 10.8LADD1037 pKa = 3.75SRR1039 pKa = 11.84TGDD1042 pKa = 3.28RR1043 pKa = 11.84TALSTANPQNGTGTPTVTFIGSTGIALLADD1073 pKa = 4.03GTTVTSISVTDD1084 pKa = 3.79FAVLQQTVVGGDD1096 pKa = 3.5LAFDD1100 pKa = 3.43WVVNPNSQFAYY1111 pKa = 10.15GVTADD1116 pKa = 3.71PTSPVTSVTVVEE1128 pKa = 4.66LTTGTITEE1136 pKa = 4.22IAVPPTSAYY1145 pKa = 10.39AIDD1148 pKa = 4.17ANGSALYY1155 pKa = 9.45TYY1157 pKa = 10.93VLDD1160 pKa = 4.33PFTFSTQVTRR1170 pKa = 11.84TPFGDD1175 pKa = 3.3AGNAISVTVPGAVAGNVPEE1194 pKa = 4.77LSWSSFDD1201 pKa = 4.84DD1202 pKa = 3.55AGNYY1206 pKa = 9.93YY1207 pKa = 9.25LTTVDD1212 pKa = 3.47VSVVAGVPTLGAVKK1226 pKa = 10.08VHH1228 pKa = 6.31VFDD1231 pKa = 4.24QAMNEE1236 pKa = 3.73IDD1238 pKa = 4.09TIVTDD1243 pKa = 3.86VPSGQVAAMTSVADD1257 pKa = 3.6GTVYY1261 pKa = 11.13LMVADD1266 pKa = 5.01PGSAQNTYY1274 pKa = 9.45TIHH1277 pKa = 6.86RR1278 pKa = 11.84VSADD1282 pKa = 3.07GSVDD1286 pKa = 3.64DD1287 pKa = 5.35LADD1290 pKa = 3.68GVGWVTADD1298 pKa = 3.43QQHH1301 pKa = 5.7MFVLRR1306 pKa = 11.84TNDD1309 pKa = 2.85GSGYY1313 pKa = 8.6NTFSVLDD1320 pKa = 4.15LTTDD1324 pKa = 3.5TQSAPISVSTSYY1336 pKa = 11.14DD1337 pKa = 3.13GEE1339 pKa = 3.96FRR1341 pKa = 11.84VFEE1344 pKa = 4.26GKK1346 pKa = 10.21YY1347 pKa = 10.18LAIAYY1352 pKa = 7.87HH1353 pKa = 6.56AGSNVYY1359 pKa = 8.75VHH1361 pKa = 6.89ILDD1364 pKa = 3.58VDD1366 pKa = 4.01GNATDD1371 pKa = 4.09FVIGADD1377 pKa = 3.65DD1378 pKa = 4.26FEE1380 pKa = 4.74GLIYY1384 pKa = 10.57LDD1386 pKa = 4.49DD1387 pKa = 3.95QFSVITSGGGYY1398 pKa = 7.32TQATLIRR1405 pKa = 11.84AA1406 pKa = 3.85
Molecular weight: 143.29 kDa
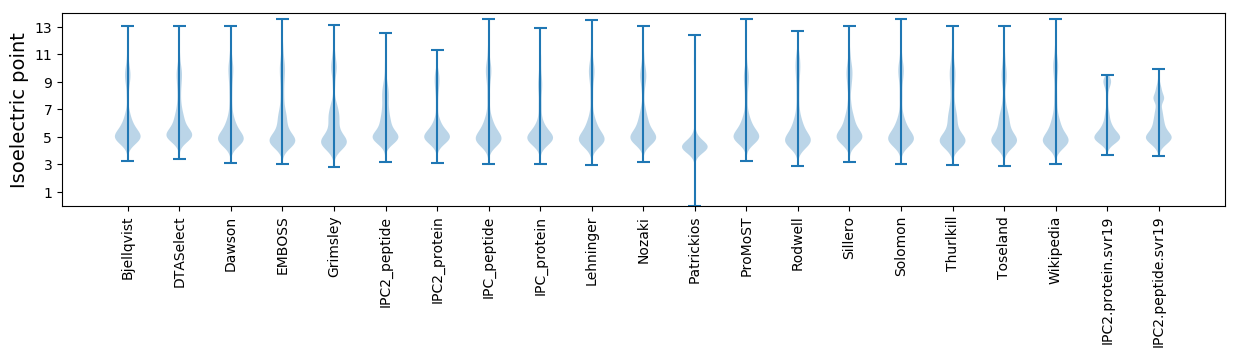
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G7GQD9|G7GQD9_9ACTN 50S ribosomal protein L15 OS=Gordonia amarae NBRC 15530 OX=1075090 GN=rplO PE=3 SV=1
MM1 pKa = 7.46TKK3 pKa = 10.27GKK5 pKa = 8.73RR6 pKa = 11.84TFQPNNRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84SRR17 pKa = 11.84VHH19 pKa = 6.33GFRR22 pKa = 11.84LRR24 pKa = 11.84MRR26 pKa = 11.84TRR28 pKa = 11.84AGRR31 pKa = 11.84AIVNSRR37 pKa = 11.84RR38 pKa = 11.84SKK40 pKa = 10.8GRR42 pKa = 11.84AKK44 pKa = 9.67LTAA47 pKa = 4.21
MM1 pKa = 7.46TKK3 pKa = 10.27GKK5 pKa = 8.73RR6 pKa = 11.84TFQPNNRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84SRR17 pKa = 11.84VHH19 pKa = 6.33GFRR22 pKa = 11.84LRR24 pKa = 11.84MRR26 pKa = 11.84TRR28 pKa = 11.84AGRR31 pKa = 11.84AIVNSRR37 pKa = 11.84RR38 pKa = 11.84SKK40 pKa = 10.8GRR42 pKa = 11.84AKK44 pKa = 9.67LTAA47 pKa = 4.21
Molecular weight: 5.54 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1599335 |
34 |
8820 |
340.9 |
36.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.748 ± 0.049 | 0.75 ± 0.011 |
6.7 ± 0.032 | 5.204 ± 0.029 |
2.968 ± 0.023 | 9.099 ± 0.03 |
2.097 ± 0.016 | 4.589 ± 0.02 |
2.417 ± 0.03 | 9.469 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.958 ± 0.016 | 2.153 ± 0.017 |
5.659 ± 0.024 | 2.739 ± 0.018 |
7.111 ± 0.036 | 5.644 ± 0.023 |
6.465 ± 0.028 | 8.721 ± 0.034 |
1.408 ± 0.013 | 2.1 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
