
Sphingomonas edaphi
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Sphingomonas
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
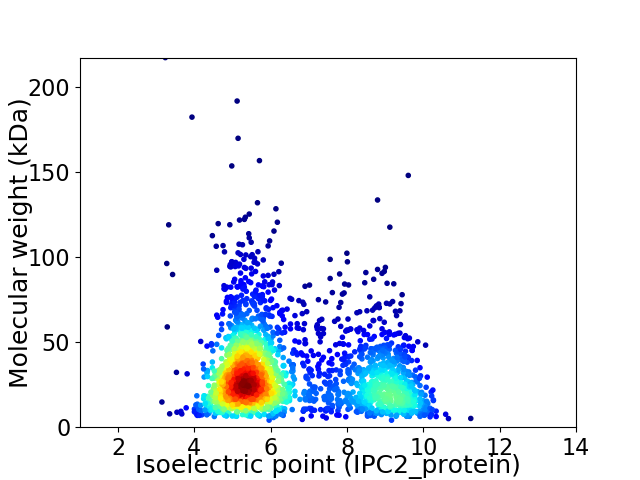
Virtual 2D-PAGE plot for 2311 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A418Q0Z4|A0A418Q0Z4_9SPHN Sigma-54-dependent Fis family transcriptional regulator OS=Sphingomonas edaphi OX=2315689 GN=D3M59_00950 PE=4 SV=1
AA1 pKa = 8.32AIEE4 pKa = 4.3ALDD7 pKa = 4.02AGEE10 pKa = 4.37TPSDD14 pKa = 3.63VFTVSVSDD22 pKa = 3.54GDD24 pKa = 4.27GPLVTQSYY32 pKa = 8.16TVNLTGADD40 pKa = 3.76DD41 pKa = 5.01NIAPIANSDD50 pKa = 4.28VIWVSNNTTVTLSNDD65 pKa = 3.02VLLGNDD71 pKa = 3.19IDD73 pKa = 4.09VDD75 pKa = 4.78GIALILTNVVVTTGLLASPVVLNANGTFTFTTGAAGGTVGTPTVVTLTYY124 pKa = 10.08STDD127 pKa = 3.44DD128 pKa = 3.72GAGGTTTGSVTVNIITTTSGVDD150 pKa = 3.59SLNLTAVPAYY160 pKa = 9.22QASFIQGLAGADD172 pKa = 3.55TLTDD176 pKa = 3.49GTGVSTLLGGDD187 pKa = 3.5GADD190 pKa = 3.54NLTGNAGNDD199 pKa = 3.63LLIGGDD205 pKa = 3.81NNDD208 pKa = 3.66VLDD211 pKa = 4.62GGSGNDD217 pKa = 3.15ILRR220 pKa = 11.84GGFGNNDD227 pKa = 3.55EE228 pKa = 4.54MNGGAGSEE236 pKa = 4.24DD237 pKa = 3.96LLDD240 pKa = 4.66FSDD243 pKa = 3.65GTAAVTFTLVQSAVFTSIANGTGGLGNNDD272 pKa = 3.33KK273 pKa = 10.63YY274 pKa = 11.4MNMEE278 pKa = 3.99GVIGTSQGDD287 pKa = 3.99TLTGSANNDD296 pKa = 2.9ILRR299 pKa = 11.84GGGGNDD305 pKa = 3.39TLNGGGGLGDD315 pKa = 5.49LIDD318 pKa = 4.71FRR320 pKa = 11.84DD321 pKa = 4.04GTAGITFTLVQGGGATVFNASAAGLGSDD349 pKa = 3.8TYY351 pKa = 11.86SNIEE355 pKa = 3.95GVIGTVFNDD364 pKa = 2.96ILTGSSGNDD373 pKa = 2.96ILRR376 pKa = 11.84GDD378 pKa = 4.26GGNDD382 pKa = 3.45VINGADD388 pKa = 3.76GDD390 pKa = 4.1DD391 pKa = 3.94TLVGGAGADD400 pKa = 3.89TITGGLGNDD409 pKa = 3.45VFVLTTPLNAVDD421 pKa = 4.64TIVDD425 pKa = 3.46YY426 pKa = 11.34SGSVGNADD434 pKa = 4.28VIDD437 pKa = 3.42ITAILNVPGAVDD449 pKa = 4.98PINDD453 pKa = 3.78GYY455 pKa = 11.61LRR457 pKa = 11.84VTTTGLLQVDD467 pKa = 4.77LDD469 pKa = 4.09GGGNNWITLANINTGVSPTVRR490 pKa = 11.84YY491 pKa = 9.59IDD493 pKa = 3.78GGVITNIVVAPVAPPIALDD512 pKa = 3.63LDD514 pKa = 3.86GDD516 pKa = 4.35GFVSFTGTDD525 pKa = 3.01GGAVFDD531 pKa = 4.27YY532 pKa = 11.01GYY534 pKa = 9.16GTVSTAWVAGNDD546 pKa = 4.39GILVRR551 pKa = 11.84DD552 pKa = 3.9ANGDD556 pKa = 3.38GHH558 pKa = 7.06ISSNEE563 pKa = 3.5IVFATTGSDD572 pKa = 3.63LEE574 pKa = 4.45GLATYY579 pKa = 7.57DD580 pKa = 3.74TNQDD584 pKa = 3.57GQLSAADD591 pKa = 3.53SQFAEE596 pKa = 4.37FGVWQDD602 pKa = 3.36ADD604 pKa = 3.4SDD606 pKa = 4.32GQVDD610 pKa = 3.58AGEE613 pKa = 4.26LQSLAAHH620 pKa = 6.87SIASISLSSDD630 pKa = 2.71GVAYY634 pKa = 10.44SAANGDD640 pKa = 3.86VQVVGTGSYY649 pKa = 9.7TRR651 pKa = 11.84TDD653 pKa = 3.01GSTGVLADD661 pKa = 3.76AVFATGNQLAANDD674 pKa = 4.03PRR676 pKa = 11.84LAPAAGNTAILAAAAAAVGLASMPAHH702 pKa = 6.56AQPADD707 pKa = 3.63SEE709 pKa = 4.65ADD711 pKa = 3.21GSFRR715 pKa = 11.84GEE717 pKa = 3.99LQEE720 pKa = 4.0TLILRR725 pKa = 11.84SHH727 pKa = 7.13ADD729 pKa = 2.9WTGLSNAASDD739 pKa = 4.21VSGLHH744 pKa = 6.76ADD746 pKa = 4.18GLASFYY752 pKa = 10.54PAMPDD757 pKa = 3.44PSQMGATPSGSNHH770 pKa = 5.81SLIGSEE776 pKa = 4.02VAVRR780 pKa = 11.84AEE782 pKa = 3.71AAFIPMEE789 pKa = 4.07MSEE792 pKa = 4.27AVHH795 pKa = 6.15VSSAPVAMAMVAMPSAEE812 pKa = 3.97LLAAAVHH819 pKa = 6.38GSGAQPHH826 pKa = 5.68GQVAAVIADD835 pKa = 4.03AFAGDD840 pKa = 3.78SHH842 pKa = 6.31ATAIDD847 pKa = 3.42ALLDD851 pKa = 3.83VVTSQHH857 pKa = 5.89NADD860 pKa = 3.58GSGGGSASLAHH871 pKa = 6.75LASAPWLAAPEE882 pKa = 3.98YY883 pKa = 10.09GQLALHH889 pKa = 5.88MQHH892 pKa = 7.15AEE894 pKa = 4.1LLVAHH899 pKa = 7.28PDD901 pKa = 3.63VLPPTT906 pKa = 4.2
AA1 pKa = 8.32AIEE4 pKa = 4.3ALDD7 pKa = 4.02AGEE10 pKa = 4.37TPSDD14 pKa = 3.63VFTVSVSDD22 pKa = 3.54GDD24 pKa = 4.27GPLVTQSYY32 pKa = 8.16TVNLTGADD40 pKa = 3.76DD41 pKa = 5.01NIAPIANSDD50 pKa = 4.28VIWVSNNTTVTLSNDD65 pKa = 3.02VLLGNDD71 pKa = 3.19IDD73 pKa = 4.09VDD75 pKa = 4.78GIALILTNVVVTTGLLASPVVLNANGTFTFTTGAAGGTVGTPTVVTLTYY124 pKa = 10.08STDD127 pKa = 3.44DD128 pKa = 3.72GAGGTTTGSVTVNIITTTSGVDD150 pKa = 3.59SLNLTAVPAYY160 pKa = 9.22QASFIQGLAGADD172 pKa = 3.55TLTDD176 pKa = 3.49GTGVSTLLGGDD187 pKa = 3.5GADD190 pKa = 3.54NLTGNAGNDD199 pKa = 3.63LLIGGDD205 pKa = 3.81NNDD208 pKa = 3.66VLDD211 pKa = 4.62GGSGNDD217 pKa = 3.15ILRR220 pKa = 11.84GGFGNNDD227 pKa = 3.55EE228 pKa = 4.54MNGGAGSEE236 pKa = 4.24DD237 pKa = 3.96LLDD240 pKa = 4.66FSDD243 pKa = 3.65GTAAVTFTLVQSAVFTSIANGTGGLGNNDD272 pKa = 3.33KK273 pKa = 10.63YY274 pKa = 11.4MNMEE278 pKa = 3.99GVIGTSQGDD287 pKa = 3.99TLTGSANNDD296 pKa = 2.9ILRR299 pKa = 11.84GGGGNDD305 pKa = 3.39TLNGGGGLGDD315 pKa = 5.49LIDD318 pKa = 4.71FRR320 pKa = 11.84DD321 pKa = 4.04GTAGITFTLVQGGGATVFNASAAGLGSDD349 pKa = 3.8TYY351 pKa = 11.86SNIEE355 pKa = 3.95GVIGTVFNDD364 pKa = 2.96ILTGSSGNDD373 pKa = 2.96ILRR376 pKa = 11.84GDD378 pKa = 4.26GGNDD382 pKa = 3.45VINGADD388 pKa = 3.76GDD390 pKa = 4.1DD391 pKa = 3.94TLVGGAGADD400 pKa = 3.89TITGGLGNDD409 pKa = 3.45VFVLTTPLNAVDD421 pKa = 4.64TIVDD425 pKa = 3.46YY426 pKa = 11.34SGSVGNADD434 pKa = 4.28VIDD437 pKa = 3.42ITAILNVPGAVDD449 pKa = 4.98PINDD453 pKa = 3.78GYY455 pKa = 11.61LRR457 pKa = 11.84VTTTGLLQVDD467 pKa = 4.77LDD469 pKa = 4.09GGGNNWITLANINTGVSPTVRR490 pKa = 11.84YY491 pKa = 9.59IDD493 pKa = 3.78GGVITNIVVAPVAPPIALDD512 pKa = 3.63LDD514 pKa = 3.86GDD516 pKa = 4.35GFVSFTGTDD525 pKa = 3.01GGAVFDD531 pKa = 4.27YY532 pKa = 11.01GYY534 pKa = 9.16GTVSTAWVAGNDD546 pKa = 4.39GILVRR551 pKa = 11.84DD552 pKa = 3.9ANGDD556 pKa = 3.38GHH558 pKa = 7.06ISSNEE563 pKa = 3.5IVFATTGSDD572 pKa = 3.63LEE574 pKa = 4.45GLATYY579 pKa = 7.57DD580 pKa = 3.74TNQDD584 pKa = 3.57GQLSAADD591 pKa = 3.53SQFAEE596 pKa = 4.37FGVWQDD602 pKa = 3.36ADD604 pKa = 3.4SDD606 pKa = 4.32GQVDD610 pKa = 3.58AGEE613 pKa = 4.26LQSLAAHH620 pKa = 6.87SIASISLSSDD630 pKa = 2.71GVAYY634 pKa = 10.44SAANGDD640 pKa = 3.86VQVVGTGSYY649 pKa = 9.7TRR651 pKa = 11.84TDD653 pKa = 3.01GSTGVLADD661 pKa = 3.76AVFATGNQLAANDD674 pKa = 4.03PRR676 pKa = 11.84LAPAAGNTAILAAAAAAVGLASMPAHH702 pKa = 6.56AQPADD707 pKa = 3.63SEE709 pKa = 4.65ADD711 pKa = 3.21GSFRR715 pKa = 11.84GEE717 pKa = 3.99LQEE720 pKa = 4.0TLILRR725 pKa = 11.84SHH727 pKa = 7.13ADD729 pKa = 2.9WTGLSNAASDD739 pKa = 4.21VSGLHH744 pKa = 6.76ADD746 pKa = 4.18GLASFYY752 pKa = 10.54PAMPDD757 pKa = 3.44PSQMGATPSGSNHH770 pKa = 5.81SLIGSEE776 pKa = 4.02VAVRR780 pKa = 11.84AEE782 pKa = 3.71AAFIPMEE789 pKa = 4.07MSEE792 pKa = 4.27AVHH795 pKa = 6.15VSSAPVAMAMVAMPSAEE812 pKa = 3.97LLAAAVHH819 pKa = 6.38GSGAQPHH826 pKa = 5.68GQVAAVIADD835 pKa = 4.03AFAGDD840 pKa = 3.78SHH842 pKa = 6.31ATAIDD847 pKa = 3.42ALLDD851 pKa = 3.83VVTSQHH857 pKa = 5.89NADD860 pKa = 3.58GSGGGSASLAHH871 pKa = 6.75LASAPWLAAPEE882 pKa = 3.98YY883 pKa = 10.09GQLALHH889 pKa = 5.88MQHH892 pKa = 7.15AEE894 pKa = 4.1LLVAHH899 pKa = 7.28PDD901 pKa = 3.63VLPPTT906 pKa = 4.2
Molecular weight: 89.79 kDa
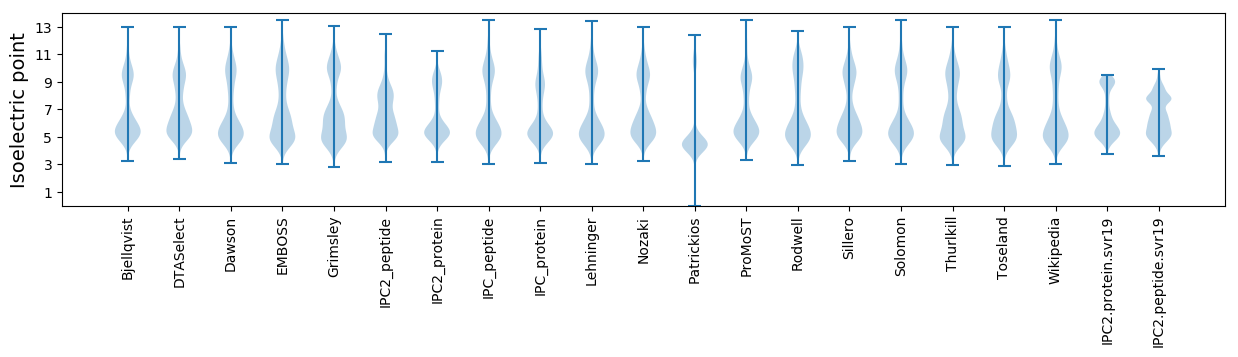
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A418Q465|A0A418Q465_9SPHN CDP-diacylglycerol--glycerol-3-phosphate 3-phosphatidyltransferase OS=Sphingomonas edaphi OX=2315689 GN=pgsA PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 8.96RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.62GFRR19 pKa = 11.84SRR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.07VLNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.02KK41 pKa = 10.61LSAA44 pKa = 4.03
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 8.96RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.62GFRR19 pKa = 11.84SRR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.07VLNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.02KK41 pKa = 10.61LSAA44 pKa = 4.03
Molecular weight: 5.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
728684 |
37 |
2143 |
315.3 |
34.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.767 ± 0.077 | 0.747 ± 0.015 |
6.017 ± 0.036 | 5.815 ± 0.051 |
3.496 ± 0.031 | 8.865 ± 0.064 |
1.903 ± 0.023 | 5.096 ± 0.031 |
3.4 ± 0.043 | 9.95 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.399 ± 0.021 | 2.632 ± 0.036 |
5.201 ± 0.039 | 3.097 ± 0.024 |
7.354 ± 0.06 | 5.53 ± 0.039 |
5.025 ± 0.038 | 7.224 ± 0.038 |
1.365 ± 0.021 | 2.118 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
