
Halostagnicola larsenii XH-48
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Natrialbales; Natrialbaceae; Halostagnicola; Halostagnicola larsenii
Average proteome isoelectric point is 4.89
Get precalculated fractions of proteins
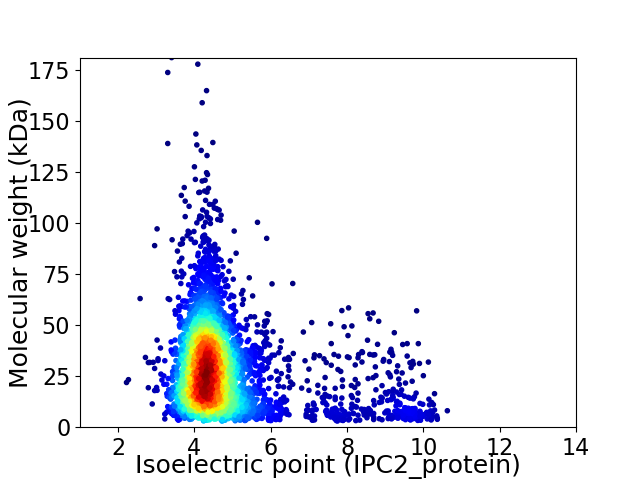
Virtual 2D-PAGE plot for 4086 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
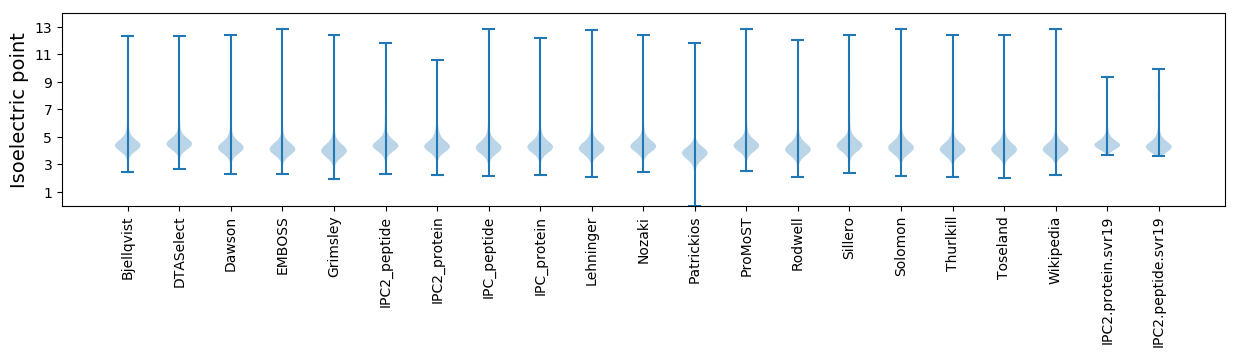
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W0JSI0|W0JSI0_9EURY Sodium:solute symporter OS=Halostagnicola larsenii XH-48 OX=797299 GN=HALLA_16980 PE=3 SV=1
MM1 pKa = 7.43GALSGLSGSAAAQSSGGSQFGLDD24 pKa = 3.68AGFADD29 pKa = 5.13TSWLDD34 pKa = 3.49DD35 pKa = 4.28DD36 pKa = 5.63VDD38 pKa = 3.71VHH40 pKa = 6.73TITEE44 pKa = 4.21PTRR47 pKa = 11.84SAVEE51 pKa = 4.09DD52 pKa = 3.71AFSASGSRR60 pKa = 11.84VVVFEE65 pKa = 3.97TSGTIDD71 pKa = 3.81LGGNDD76 pKa = 3.99LSISEE81 pKa = 4.64DD82 pKa = 3.94YY83 pKa = 10.95CWVAGQTAPSPGITFINGQVQIDD106 pKa = 3.72ADD108 pKa = 3.89NCVVQHH114 pKa = 5.3IRR116 pKa = 11.84SRR118 pKa = 11.84IGPGSDD124 pKa = 4.29GSIQSNDD131 pKa = 3.4SFNTADD137 pKa = 3.47DD138 pKa = 3.92TQNNVVDD145 pKa = 4.49HH146 pKa = 6.64VSASWGTDD154 pKa = 2.56EE155 pKa = 4.91CLSVGYY161 pKa = 8.02DD162 pKa = 3.51TQDD165 pKa = 3.21TTVSNCLIYY174 pKa = 10.61EE175 pKa = 4.4GLYY178 pKa = 10.68DD179 pKa = 4.03PYY181 pKa = 11.4GDD183 pKa = 4.53EE184 pKa = 4.45SDD186 pKa = 4.34HH187 pKa = 7.17NYY189 pKa = 10.63GSLIGDD195 pKa = 4.09GASNVTLAGNVWAKK209 pKa = 10.16VRR211 pKa = 11.84GRR213 pKa = 11.84APRR216 pKa = 11.84LKK218 pKa = 10.39SDD220 pKa = 3.45TEE222 pKa = 4.21TAVVNNLLYY231 pKa = 10.89FFDD234 pKa = 4.18EE235 pKa = 4.43SANADD240 pKa = 3.47SSAVTSFVGNAAICADD256 pKa = 4.48DD257 pKa = 5.56DD258 pKa = 4.28DD259 pKa = 7.06AILEE263 pKa = 4.33GSPTAYY269 pKa = 10.31HH270 pKa = 7.33ADD272 pKa = 3.71NIAYY276 pKa = 7.95DD277 pKa = 4.11PPMEE281 pKa = 5.39DD282 pKa = 3.97DD283 pKa = 4.01QPIAEE288 pKa = 4.7PEE290 pKa = 4.57SVSSPPLWPSGLSEE304 pKa = 3.82MAANSVEE311 pKa = 4.05DD312 pKa = 4.21HH313 pKa = 6.45NLANAGARR321 pKa = 11.84PADD324 pKa = 4.04RR325 pKa = 11.84TAHH328 pKa = 5.9DD329 pKa = 3.4QRR331 pKa = 11.84IVQEE335 pKa = 3.79IADD338 pKa = 4.61RR339 pKa = 11.84DD340 pKa = 3.86GLDD343 pKa = 3.98YY344 pKa = 11.24LDD346 pKa = 4.62SPYY349 pKa = 10.66DD350 pKa = 3.69YY351 pKa = 10.64WVGAPDD357 pKa = 4.87DD358 pKa = 3.89VGGYY362 pKa = 9.51PDD364 pKa = 5.32LPVNTHH370 pKa = 5.38SLEE373 pKa = 4.27VPDD376 pKa = 3.8SDD378 pKa = 4.43IRR380 pKa = 11.84GWLAGWAQSVEE391 pKa = 4.13
MM1 pKa = 7.43GALSGLSGSAAAQSSGGSQFGLDD24 pKa = 3.68AGFADD29 pKa = 5.13TSWLDD34 pKa = 3.49DD35 pKa = 4.28DD36 pKa = 5.63VDD38 pKa = 3.71VHH40 pKa = 6.73TITEE44 pKa = 4.21PTRR47 pKa = 11.84SAVEE51 pKa = 4.09DD52 pKa = 3.71AFSASGSRR60 pKa = 11.84VVVFEE65 pKa = 3.97TSGTIDD71 pKa = 3.81LGGNDD76 pKa = 3.99LSISEE81 pKa = 4.64DD82 pKa = 3.94YY83 pKa = 10.95CWVAGQTAPSPGITFINGQVQIDD106 pKa = 3.72ADD108 pKa = 3.89NCVVQHH114 pKa = 5.3IRR116 pKa = 11.84SRR118 pKa = 11.84IGPGSDD124 pKa = 4.29GSIQSNDD131 pKa = 3.4SFNTADD137 pKa = 3.47DD138 pKa = 3.92TQNNVVDD145 pKa = 4.49HH146 pKa = 6.64VSASWGTDD154 pKa = 2.56EE155 pKa = 4.91CLSVGYY161 pKa = 8.02DD162 pKa = 3.51TQDD165 pKa = 3.21TTVSNCLIYY174 pKa = 10.61EE175 pKa = 4.4GLYY178 pKa = 10.68DD179 pKa = 4.03PYY181 pKa = 11.4GDD183 pKa = 4.53EE184 pKa = 4.45SDD186 pKa = 4.34HH187 pKa = 7.17NYY189 pKa = 10.63GSLIGDD195 pKa = 4.09GASNVTLAGNVWAKK209 pKa = 10.16VRR211 pKa = 11.84GRR213 pKa = 11.84APRR216 pKa = 11.84LKK218 pKa = 10.39SDD220 pKa = 3.45TEE222 pKa = 4.21TAVVNNLLYY231 pKa = 10.89FFDD234 pKa = 4.18EE235 pKa = 4.43SANADD240 pKa = 3.47SSAVTSFVGNAAICADD256 pKa = 4.48DD257 pKa = 5.56DD258 pKa = 4.28DD259 pKa = 7.06AILEE263 pKa = 4.33GSPTAYY269 pKa = 10.31HH270 pKa = 7.33ADD272 pKa = 3.71NIAYY276 pKa = 7.95DD277 pKa = 4.11PPMEE281 pKa = 5.39DD282 pKa = 3.97DD283 pKa = 4.01QPIAEE288 pKa = 4.7PEE290 pKa = 4.57SVSSPPLWPSGLSEE304 pKa = 3.82MAANSVEE311 pKa = 4.05DD312 pKa = 4.21HH313 pKa = 6.45NLANAGARR321 pKa = 11.84PADD324 pKa = 4.04RR325 pKa = 11.84TAHH328 pKa = 5.9DD329 pKa = 3.4QRR331 pKa = 11.84IVQEE335 pKa = 3.79IADD338 pKa = 4.61RR339 pKa = 11.84DD340 pKa = 3.86GLDD343 pKa = 3.98YY344 pKa = 11.24LDD346 pKa = 4.62SPYY349 pKa = 10.66DD350 pKa = 3.69YY351 pKa = 10.64WVGAPDD357 pKa = 4.87DD358 pKa = 3.89VGGYY362 pKa = 9.51PDD364 pKa = 5.32LPVNTHH370 pKa = 5.38SLEE373 pKa = 4.27VPDD376 pKa = 3.8SDD378 pKa = 4.43IRR380 pKa = 11.84GWLAGWAQSVEE391 pKa = 4.13
Molecular weight: 41.28 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W0JYJ7|W0JYJ7_9EURY Acyl-CoA dehydrogenase OS=Halostagnicola larsenii XH-48 OX=797299 GN=HALLA_01905 PE=3 SV=1
MM1 pKa = 7.77RR2 pKa = 11.84PLIKK6 pKa = 10.13HH7 pKa = 6.0RR8 pKa = 11.84EE9 pKa = 3.81FRR11 pKa = 11.84PIDD14 pKa = 3.56HH15 pKa = 6.86AHH17 pKa = 6.44NARR20 pKa = 11.84IDD22 pKa = 3.49GTLYY26 pKa = 10.37GQSALSVTVFSVIKK40 pKa = 9.23RR41 pKa = 11.84TLGVAEE47 pKa = 4.94RR48 pKa = 11.84ARR50 pKa = 11.84SWYY53 pKa = 9.98RR54 pKa = 11.84EE55 pKa = 3.69LRR57 pKa = 11.84EE58 pKa = 4.06IVLMCAVYY66 pKa = 10.13NIKK69 pKa = 10.48RR70 pKa = 11.84AVSS73 pKa = 3.29
MM1 pKa = 7.77RR2 pKa = 11.84PLIKK6 pKa = 10.13HH7 pKa = 6.0RR8 pKa = 11.84EE9 pKa = 3.81FRR11 pKa = 11.84PIDD14 pKa = 3.56HH15 pKa = 6.86AHH17 pKa = 6.44NARR20 pKa = 11.84IDD22 pKa = 3.49GTLYY26 pKa = 10.37GQSALSVTVFSVIKK40 pKa = 9.23RR41 pKa = 11.84TLGVAEE47 pKa = 4.94RR48 pKa = 11.84ARR50 pKa = 11.84SWYY53 pKa = 9.98RR54 pKa = 11.84EE55 pKa = 3.69LRR57 pKa = 11.84EE58 pKa = 4.06IVLMCAVYY66 pKa = 10.13NIKK69 pKa = 10.48RR70 pKa = 11.84AVSS73 pKa = 3.29
Molecular weight: 8.47 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1126012 |
29 |
1757 |
275.6 |
30.08 |
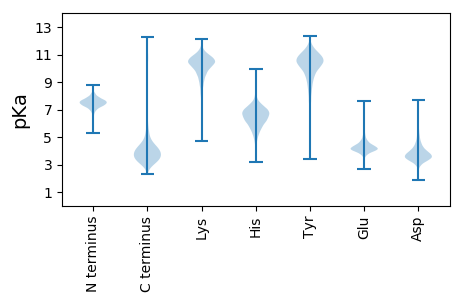
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.931 ± 0.05 | 0.743 ± 0.012 |
8.263 ± 0.05 | 9.229 ± 0.051 |
3.381 ± 0.033 | 8.313 ± 0.041 |
1.999 ± 0.018 | 4.844 ± 0.028 |
1.873 ± 0.021 | 8.743 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.841 ± 0.016 | 2.566 ± 0.022 |
4.528 ± 0.028 | 2.631 ± 0.022 |
6.147 ± 0.038 | 6.054 ± 0.033 |
6.576 ± 0.031 | 8.457 ± 0.036 |
1.13 ± 0.016 | 2.751 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
