
Lachancea mirantina
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Saccharomycotina; Saccharomycetes; Saccharomycetales; Saccharomycetaceae; Lachancea
Average proteome isoelectric point is 6.56
Get precalculated fractions of proteins
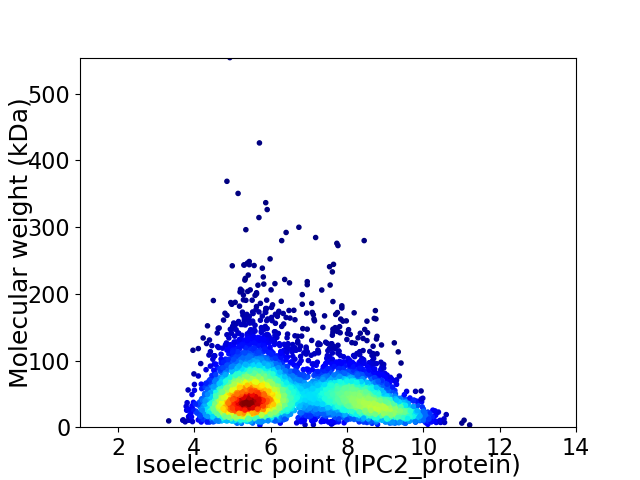
Virtual 2D-PAGE plot for 5053 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G4J9J0|A0A1G4J9J0_9SACH LAMI_0D02168g1_1 OS=Lachancea mirantina OX=1230905 GN=LAMI_0D02168G PE=4 SV=1
MM1 pKa = 7.62LSSQQIVALTALVGLAQASWWDD23 pKa = 3.54GFLNPNATSSSSTAPAVVSSSQATDD48 pKa = 3.38LASASSQSSAVASTAPAIQPYY69 pKa = 9.85ISSSSQGAQATLSPISSEE87 pKa = 4.08TSSEE91 pKa = 4.05GGQTTLSPTSSDD103 pKa = 3.27VSSEE107 pKa = 4.37DD108 pKa = 3.52GSQSFQAYY116 pKa = 9.81LSSIMQATQTQGSSAFTVYY135 pKa = 10.1PSSSSYY141 pKa = 11.19SSPQDD146 pKa = 3.37TTTLAPTFSADD157 pKa = 3.15SFTSATSTTSDD168 pKa = 2.71WNTFSSSEE176 pKa = 3.93QSLTPSSEE184 pKa = 4.36SKK186 pKa = 11.04LEE188 pKa = 4.15SQLSQSASFSTSAWGFSNTRR208 pKa = 11.84STPVEE213 pKa = 3.89KK214 pKa = 10.81SFSSSSALASEE225 pKa = 4.66SATLATPQNTILDD238 pKa = 4.03SSEE241 pKa = 4.36SPTASEE247 pKa = 4.69PTTSAPKK254 pKa = 7.83WTSSASPEE262 pKa = 4.51SVTSAAPDD270 pKa = 3.27ATKK273 pKa = 10.49PILSEE278 pKa = 4.37SASSTGPIPSSEE290 pKa = 4.13ASWLSSEE297 pKa = 5.09QYY299 pKa = 9.34TATQRR304 pKa = 11.84EE305 pKa = 4.68VTTASATPSSANVAASSVEE324 pKa = 3.93IQSEE328 pKa = 4.52VQSASSFGSFSASEE342 pKa = 4.14TSSTEE347 pKa = 3.84VDD349 pKa = 3.0SSTALQAKK357 pKa = 9.81SSPQTSTWPASTAVSLSIVPTSSSEE382 pKa = 4.29SQVSSALSVTSSSLTPASAAPSSSEE407 pKa = 3.52FSIEE411 pKa = 3.86QSSSQFEE418 pKa = 4.36DD419 pKa = 3.54TSITGVDD426 pKa = 3.87WNTVSYY432 pKa = 9.2PASSSAADD440 pKa = 3.68VPSSTQYY447 pKa = 11.2SSQQAQPTSQSPLWSSSGAQKK468 pKa = 9.71TYY470 pKa = 10.79SYY472 pKa = 11.22VPSTSSSSYY481 pKa = 10.84DD482 pKa = 3.17SSTAEE487 pKa = 3.99SWASTPASQSSASDD501 pKa = 2.88ISNYY505 pKa = 10.44AEE507 pKa = 3.99TWQTASSSASPSSYY521 pKa = 10.72LPSSSNYY528 pKa = 9.93AGSTSSYY535 pKa = 10.47PYY537 pKa = 9.15LTSSQIYY544 pKa = 7.97ATSSSDD550 pKa = 3.24SYY552 pKa = 11.78EE553 pKa = 4.15SFPPTSSLASSEE565 pKa = 4.06YY566 pKa = 9.61STLSSEE572 pKa = 4.51ASLLSTSSQSAFSVTPSASSEE593 pKa = 4.15SSFTSTVPAPSSLSASSFVTDD614 pKa = 3.82DD615 pKa = 4.11FSRR618 pKa = 11.84SSSSLPTSSSAATQSTTSLMVSSSQSTSDD647 pKa = 3.3STSIASVSSFPEE659 pKa = 4.12SSVSTPAPSSLVTVSSYY676 pKa = 11.45ASEE679 pKa = 4.2NATSSALIASLSSSPSASITSTAEE703 pKa = 3.56SSEE706 pKa = 3.97FSSALFTSSYY716 pKa = 10.25NQTDD720 pKa = 3.59TGSLASSASTEE731 pKa = 4.32SVSEE735 pKa = 4.27SLFSATVSATSSFEE749 pKa = 4.28SSVSTPIAIPSSATSALTTGAPEE772 pKa = 3.85QVTSSEE778 pKa = 4.31VAQNSSSLGSSYY790 pKa = 10.99TSSTSNWLPTAIITAGGDD808 pKa = 3.65TQSTGSGAAASGGQQATQTLPQAIAAATAVSQPKK842 pKa = 10.15DD843 pKa = 3.48YY844 pKa = 10.49TLITIGFKK852 pKa = 10.44QEE854 pKa = 3.5LNYY857 pKa = 10.4PFVVKK862 pKa = 10.83NSLTSAQIFAYY873 pKa = 10.44LPGVLNLPFEE883 pKa = 4.98DD884 pKa = 5.24SFDD887 pKa = 3.91QIQVVQLSPLNVKK900 pKa = 9.43GRR902 pKa = 11.84SYY904 pKa = 11.19LSTVAEE910 pKa = 4.7VYY912 pKa = 10.45FPSSEE917 pKa = 3.73VDD919 pKa = 3.1ALQEE923 pKa = 3.81LVQNSEE929 pKa = 4.08SKK931 pKa = 10.88LYY933 pKa = 10.92LSANSALAGLIDD945 pKa = 3.63TTIPLTGLVQSSDD958 pKa = 3.26GSSDD962 pKa = 3.48DD963 pKa = 3.85TSSQSSTSQSNSEE976 pKa = 4.25GNTDD980 pKa = 3.03STGGSRR986 pKa = 11.84GRR988 pKa = 11.84IAGTLDD994 pKa = 2.99VTYY997 pKa = 10.6NASASSKK1004 pKa = 9.91GQPGNNGKK1012 pKa = 9.55LVGLIIGVVVGAIVYY1027 pKa = 8.94LAFMVVGYY1035 pKa = 10.48RR1036 pKa = 11.84FFVLKK1041 pKa = 10.36RR1042 pKa = 11.84KK1043 pKa = 8.29QQAHH1047 pKa = 6.22SNDD1050 pKa = 3.8LGDD1053 pKa = 4.17GQSLGSSTDD1062 pKa = 3.16HH1063 pKa = 7.24GSYY1066 pKa = 10.64LYY1068 pKa = 10.35DD1069 pKa = 3.45EE1070 pKa = 5.41KK1071 pKa = 11.58EE1072 pKa = 4.13NGDD1075 pKa = 3.89QVSLSPSMRR1084 pKa = 11.84INNWMRR1090 pKa = 11.84HH1091 pKa = 3.8NHH1093 pKa = 5.19YY1094 pKa = 9.59EE1095 pKa = 3.99YY1096 pKa = 10.89SGEE1099 pKa = 4.09GQQNLGEE1106 pKa = 4.17QHH1108 pKa = 6.64SSQPKK1113 pKa = 7.83ISRR1116 pKa = 11.84PIATEE1121 pKa = 4.26NSLGWNDD1128 pKa = 3.22VV1129 pKa = 3.19
MM1 pKa = 7.62LSSQQIVALTALVGLAQASWWDD23 pKa = 3.54GFLNPNATSSSSTAPAVVSSSQATDD48 pKa = 3.38LASASSQSSAVASTAPAIQPYY69 pKa = 9.85ISSSSQGAQATLSPISSEE87 pKa = 4.08TSSEE91 pKa = 4.05GGQTTLSPTSSDD103 pKa = 3.27VSSEE107 pKa = 4.37DD108 pKa = 3.52GSQSFQAYY116 pKa = 9.81LSSIMQATQTQGSSAFTVYY135 pKa = 10.1PSSSSYY141 pKa = 11.19SSPQDD146 pKa = 3.37TTTLAPTFSADD157 pKa = 3.15SFTSATSTTSDD168 pKa = 2.71WNTFSSSEE176 pKa = 3.93QSLTPSSEE184 pKa = 4.36SKK186 pKa = 11.04LEE188 pKa = 4.15SQLSQSASFSTSAWGFSNTRR208 pKa = 11.84STPVEE213 pKa = 3.89KK214 pKa = 10.81SFSSSSALASEE225 pKa = 4.66SATLATPQNTILDD238 pKa = 4.03SSEE241 pKa = 4.36SPTASEE247 pKa = 4.69PTTSAPKK254 pKa = 7.83WTSSASPEE262 pKa = 4.51SVTSAAPDD270 pKa = 3.27ATKK273 pKa = 10.49PILSEE278 pKa = 4.37SASSTGPIPSSEE290 pKa = 4.13ASWLSSEE297 pKa = 5.09QYY299 pKa = 9.34TATQRR304 pKa = 11.84EE305 pKa = 4.68VTTASATPSSANVAASSVEE324 pKa = 3.93IQSEE328 pKa = 4.52VQSASSFGSFSASEE342 pKa = 4.14TSSTEE347 pKa = 3.84VDD349 pKa = 3.0SSTALQAKK357 pKa = 9.81SSPQTSTWPASTAVSLSIVPTSSSEE382 pKa = 4.29SQVSSALSVTSSSLTPASAAPSSSEE407 pKa = 3.52FSIEE411 pKa = 3.86QSSSQFEE418 pKa = 4.36DD419 pKa = 3.54TSITGVDD426 pKa = 3.87WNTVSYY432 pKa = 9.2PASSSAADD440 pKa = 3.68VPSSTQYY447 pKa = 11.2SSQQAQPTSQSPLWSSSGAQKK468 pKa = 9.71TYY470 pKa = 10.79SYY472 pKa = 11.22VPSTSSSSYY481 pKa = 10.84DD482 pKa = 3.17SSTAEE487 pKa = 3.99SWASTPASQSSASDD501 pKa = 2.88ISNYY505 pKa = 10.44AEE507 pKa = 3.99TWQTASSSASPSSYY521 pKa = 10.72LPSSSNYY528 pKa = 9.93AGSTSSYY535 pKa = 10.47PYY537 pKa = 9.15LTSSQIYY544 pKa = 7.97ATSSSDD550 pKa = 3.24SYY552 pKa = 11.78EE553 pKa = 4.15SFPPTSSLASSEE565 pKa = 4.06YY566 pKa = 9.61STLSSEE572 pKa = 4.51ASLLSTSSQSAFSVTPSASSEE593 pKa = 4.15SSFTSTVPAPSSLSASSFVTDD614 pKa = 3.82DD615 pKa = 4.11FSRR618 pKa = 11.84SSSSLPTSSSAATQSTTSLMVSSSQSTSDD647 pKa = 3.3STSIASVSSFPEE659 pKa = 4.12SSVSTPAPSSLVTVSSYY676 pKa = 11.45ASEE679 pKa = 4.2NATSSALIASLSSSPSASITSTAEE703 pKa = 3.56SSEE706 pKa = 3.97FSSALFTSSYY716 pKa = 10.25NQTDD720 pKa = 3.59TGSLASSASTEE731 pKa = 4.32SVSEE735 pKa = 4.27SLFSATVSATSSFEE749 pKa = 4.28SSVSTPIAIPSSATSALTTGAPEE772 pKa = 3.85QVTSSEE778 pKa = 4.31VAQNSSSLGSSYY790 pKa = 10.99TSSTSNWLPTAIITAGGDD808 pKa = 3.65TQSTGSGAAASGGQQATQTLPQAIAAATAVSQPKK842 pKa = 10.15DD843 pKa = 3.48YY844 pKa = 10.49TLITIGFKK852 pKa = 10.44QEE854 pKa = 3.5LNYY857 pKa = 10.4PFVVKK862 pKa = 10.83NSLTSAQIFAYY873 pKa = 10.44LPGVLNLPFEE883 pKa = 4.98DD884 pKa = 5.24SFDD887 pKa = 3.91QIQVVQLSPLNVKK900 pKa = 9.43GRR902 pKa = 11.84SYY904 pKa = 11.19LSTVAEE910 pKa = 4.7VYY912 pKa = 10.45FPSSEE917 pKa = 3.73VDD919 pKa = 3.1ALQEE923 pKa = 3.81LVQNSEE929 pKa = 4.08SKK931 pKa = 10.88LYY933 pKa = 10.92LSANSALAGLIDD945 pKa = 3.63TTIPLTGLVQSSDD958 pKa = 3.26GSSDD962 pKa = 3.48DD963 pKa = 3.85TSSQSSTSQSNSEE976 pKa = 4.25GNTDD980 pKa = 3.03STGGSRR986 pKa = 11.84GRR988 pKa = 11.84IAGTLDD994 pKa = 2.99VTYY997 pKa = 10.6NASASSKK1004 pKa = 9.91GQPGNNGKK1012 pKa = 9.55LVGLIIGVVVGAIVYY1027 pKa = 8.94LAFMVVGYY1035 pKa = 10.48RR1036 pKa = 11.84FFVLKK1041 pKa = 10.36RR1042 pKa = 11.84KK1043 pKa = 8.29QQAHH1047 pKa = 6.22SNDD1050 pKa = 3.8LGDD1053 pKa = 4.17GQSLGSSTDD1062 pKa = 3.16HH1063 pKa = 7.24GSYY1066 pKa = 10.64LYY1068 pKa = 10.35DD1069 pKa = 3.45EE1070 pKa = 5.41KK1071 pKa = 11.58EE1072 pKa = 4.13NGDD1075 pKa = 3.89QVSLSPSMRR1084 pKa = 11.84INNWMRR1090 pKa = 11.84HH1091 pKa = 3.8NHH1093 pKa = 5.19YY1094 pKa = 9.59EE1095 pKa = 3.99YY1096 pKa = 10.89SGEE1099 pKa = 4.09GQQNLGEE1106 pKa = 4.17QHH1108 pKa = 6.64SSQPKK1113 pKa = 7.83ISRR1116 pKa = 11.84PIATEE1121 pKa = 4.26NSLGWNDD1128 pKa = 3.22VV1129 pKa = 3.19
Molecular weight: 115.39 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G4JQ71|A0A1G4JQ71_9SACH Enhancer of translation termination 1 OS=Lachancea mirantina OX=1230905 GN=LAMI_0E12618G PE=3 SV=1
MM1 pKa = 7.1RR2 pKa = 11.84AKK4 pKa = 9.11WRR6 pKa = 11.84KK7 pKa = 9.07KK8 pKa = 6.93RR9 pKa = 11.84TRR11 pKa = 11.84RR12 pKa = 11.84LKK14 pKa = 9.48RR15 pKa = 11.84KK16 pKa = 8.41RR17 pKa = 11.84RR18 pKa = 11.84KK19 pKa = 9.19VRR21 pKa = 11.84ARR23 pKa = 11.84SKK25 pKa = 10.98
MM1 pKa = 7.1RR2 pKa = 11.84AKK4 pKa = 9.11WRR6 pKa = 11.84KK7 pKa = 9.07KK8 pKa = 6.93RR9 pKa = 11.84TRR11 pKa = 11.84RR12 pKa = 11.84LKK14 pKa = 9.48RR15 pKa = 11.84KK16 pKa = 8.41RR17 pKa = 11.84RR18 pKa = 11.84KK19 pKa = 9.19VRR21 pKa = 11.84ARR23 pKa = 11.84SKK25 pKa = 10.98
Molecular weight: 3.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2449895 |
25 |
4908 |
484.8 |
54.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.917 ± 0.029 | 1.346 ± 0.012 |
5.783 ± 0.023 | 6.632 ± 0.029 |
4.502 ± 0.02 | 5.676 ± 0.027 |
2.181 ± 0.012 | 5.505 ± 0.022 |
6.408 ± 0.033 | 9.887 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.048 ± 0.011 | 4.691 ± 0.017 |
4.437 ± 0.026 | 4.135 ± 0.028 |
5.253 ± 0.022 | 8.715 ± 0.04 |
5.45 ± 0.025 | 6.277 ± 0.023 |
1.101 ± 0.011 | 3.057 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
