
Anaeromicrobium sediminis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Clostridiaceae; Anaeromicrobium
Average proteome isoelectric point is 6.55
Get precalculated fractions of proteins
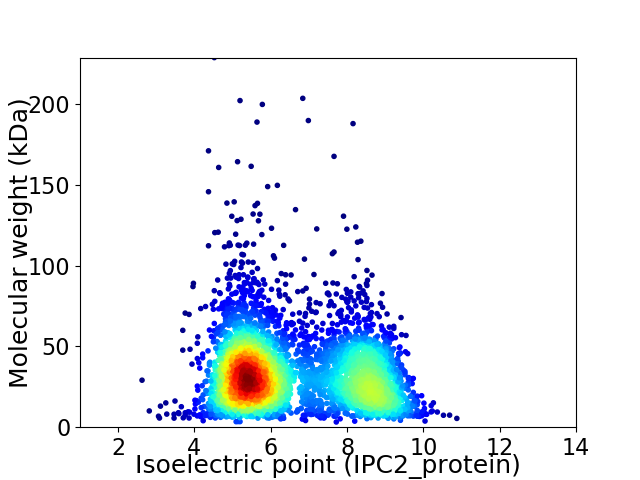
Virtual 2D-PAGE plot for 4258 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A267MKM4|A0A267MKM4_9CLOT Pyridoxamine 5'-phosphate oxidase OS=Anaeromicrobium sediminis OX=1478221 GN=CCE28_07155 PE=4 SV=1
MM1 pKa = 7.13MSAMYY6 pKa = 10.3SGVSGMQAHH15 pKa = 5.17QVKK18 pKa = 10.64LDD20 pKa = 3.79VISNNIANINTVGYY34 pKa = 9.66KK35 pKa = 10.06AQTVSFEE42 pKa = 4.18EE43 pKa = 5.4VYY45 pKa = 10.99SQTLNGATAPSDD57 pKa = 3.5SGRR60 pKa = 11.84GGTNPMQLGHH70 pKa = 6.48GVTVSSINTINTQGSMQLTGGTTDD94 pKa = 3.98FAIDD98 pKa = 3.32GDD100 pKa = 4.54GYY102 pKa = 10.97FIVQDD107 pKa = 3.63PSGAYY112 pKa = 8.61MFTRR116 pKa = 11.84AGNFGLDD123 pKa = 3.12TDD125 pKa = 4.66GNLVTADD132 pKa = 3.5GLYY135 pKa = 8.86VCGWQEE141 pKa = 4.34YY142 pKa = 8.77EE143 pKa = 4.22ALPDD147 pKa = 3.37GTYY150 pKa = 10.98VFDD153 pKa = 3.7TTKK156 pKa = 10.85DD157 pKa = 3.54PEE159 pKa = 4.99PINIFSDD166 pKa = 3.96EE167 pKa = 4.11YY168 pKa = 11.58NKK170 pKa = 10.73NKK172 pKa = 10.53NNIAPKK178 pKa = 8.48ATEE181 pKa = 3.78NVVLNGNLDD190 pKa = 3.43ASEE193 pKa = 4.31SPKK196 pKa = 10.87GSGTSDD202 pKa = 2.4IGTPPEE208 pKa = 4.04TADD211 pKa = 3.11FTMPFTAYY219 pKa = 10.43DD220 pKa = 3.74DD221 pKa = 4.47LGNDD225 pKa = 3.48YY226 pKa = 10.93EE227 pKa = 4.61VDD229 pKa = 3.81VNFTKK234 pKa = 10.64CYY236 pKa = 10.7VDD238 pKa = 6.13DD239 pKa = 4.96SDD241 pKa = 4.77PDD243 pKa = 3.74NPVTTWYY250 pKa = 9.96WEE252 pKa = 4.13MPSDD256 pKa = 4.3GSSSSSGYY264 pKa = 10.36IKK266 pKa = 10.32FDD268 pKa = 3.2KK269 pKa = 10.7DD270 pKa = 3.47GNIVEE275 pKa = 4.64EE276 pKa = 5.03DD277 pKa = 3.69GFDD280 pKa = 3.27TKK282 pKa = 11.21VDD284 pKa = 3.76VEE286 pKa = 4.38VTPDD290 pKa = 3.06SSTGTGSFTMTIDD303 pKa = 3.85FEE305 pKa = 4.78SVSMYY310 pKa = 10.62DD311 pKa = 3.62AEE313 pKa = 5.07SSVNPLNVDD322 pKa = 4.24GYY324 pKa = 10.36PSAEE328 pKa = 4.19LNDD331 pKa = 4.38FYY333 pKa = 10.96IGADD337 pKa = 3.6GVIMGVYY344 pKa = 10.42SNGQQQPLGMMSLATFSNPAGLEE367 pKa = 3.78RR368 pKa = 11.84VGNNLFMATANSGDD382 pKa = 3.69FNQAYY387 pKa = 10.31APGTQGAGTLSVGFLEE403 pKa = 4.5MSNVDD408 pKa = 3.63LSSEE412 pKa = 4.0FTEE415 pKa = 4.07MVIAQRR421 pKa = 11.84GFQANSRR428 pKa = 11.84IITTADD434 pKa = 2.97EE435 pKa = 4.78MIQEE439 pKa = 5.12LINLKK444 pKa = 10.3RR445 pKa = 3.56
MM1 pKa = 7.13MSAMYY6 pKa = 10.3SGVSGMQAHH15 pKa = 5.17QVKK18 pKa = 10.64LDD20 pKa = 3.79VISNNIANINTVGYY34 pKa = 9.66KK35 pKa = 10.06AQTVSFEE42 pKa = 4.18EE43 pKa = 5.4VYY45 pKa = 10.99SQTLNGATAPSDD57 pKa = 3.5SGRR60 pKa = 11.84GGTNPMQLGHH70 pKa = 6.48GVTVSSINTINTQGSMQLTGGTTDD94 pKa = 3.98FAIDD98 pKa = 3.32GDD100 pKa = 4.54GYY102 pKa = 10.97FIVQDD107 pKa = 3.63PSGAYY112 pKa = 8.61MFTRR116 pKa = 11.84AGNFGLDD123 pKa = 3.12TDD125 pKa = 4.66GNLVTADD132 pKa = 3.5GLYY135 pKa = 8.86VCGWQEE141 pKa = 4.34YY142 pKa = 8.77EE143 pKa = 4.22ALPDD147 pKa = 3.37GTYY150 pKa = 10.98VFDD153 pKa = 3.7TTKK156 pKa = 10.85DD157 pKa = 3.54PEE159 pKa = 4.99PINIFSDD166 pKa = 3.96EE167 pKa = 4.11YY168 pKa = 11.58NKK170 pKa = 10.73NKK172 pKa = 10.53NNIAPKK178 pKa = 8.48ATEE181 pKa = 3.78NVVLNGNLDD190 pKa = 3.43ASEE193 pKa = 4.31SPKK196 pKa = 10.87GSGTSDD202 pKa = 2.4IGTPPEE208 pKa = 4.04TADD211 pKa = 3.11FTMPFTAYY219 pKa = 10.43DD220 pKa = 3.74DD221 pKa = 4.47LGNDD225 pKa = 3.48YY226 pKa = 10.93EE227 pKa = 4.61VDD229 pKa = 3.81VNFTKK234 pKa = 10.64CYY236 pKa = 10.7VDD238 pKa = 6.13DD239 pKa = 4.96SDD241 pKa = 4.77PDD243 pKa = 3.74NPVTTWYY250 pKa = 9.96WEE252 pKa = 4.13MPSDD256 pKa = 4.3GSSSSSGYY264 pKa = 10.36IKK266 pKa = 10.32FDD268 pKa = 3.2KK269 pKa = 10.7DD270 pKa = 3.47GNIVEE275 pKa = 4.64EE276 pKa = 5.03DD277 pKa = 3.69GFDD280 pKa = 3.27TKK282 pKa = 11.21VDD284 pKa = 3.76VEE286 pKa = 4.38VTPDD290 pKa = 3.06SSTGTGSFTMTIDD303 pKa = 3.85FEE305 pKa = 4.78SVSMYY310 pKa = 10.62DD311 pKa = 3.62AEE313 pKa = 5.07SSVNPLNVDD322 pKa = 4.24GYY324 pKa = 10.36PSAEE328 pKa = 4.19LNDD331 pKa = 4.38FYY333 pKa = 10.96IGADD337 pKa = 3.6GVIMGVYY344 pKa = 10.42SNGQQQPLGMMSLATFSNPAGLEE367 pKa = 3.78RR368 pKa = 11.84VGNNLFMATANSGDD382 pKa = 3.69FNQAYY387 pKa = 10.31APGTQGAGTLSVGFLEE403 pKa = 4.5MSNVDD408 pKa = 3.63LSSEE412 pKa = 4.0FTEE415 pKa = 4.07MVIAQRR421 pKa = 11.84GFQANSRR428 pKa = 11.84IITTADD434 pKa = 2.97EE435 pKa = 4.78MIQEE439 pKa = 5.12LINLKK444 pKa = 10.3RR445 pKa = 3.56
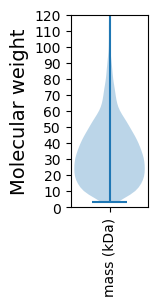
Molecular weight: 47.68 kDa
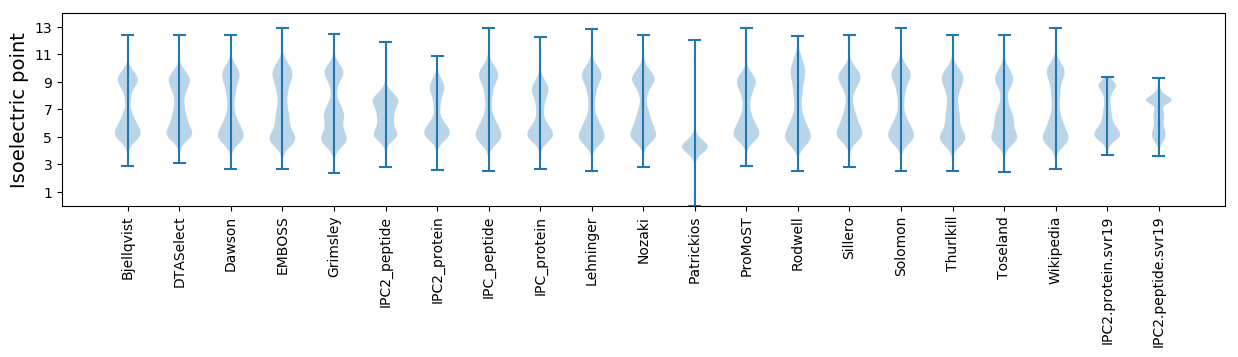
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A267MF73|A0A267MF73_9CLOT Uncharacterized protein OS=Anaeromicrobium sediminis OX=1478221 GN=CCE28_16485 PE=3 SV=1
MM1 pKa = 7.35ARR3 pKa = 11.84KK4 pKa = 9.59ALVVKK9 pKa = 7.44QQRR12 pKa = 11.84KK13 pKa = 6.5QKK15 pKa = 10.41YY16 pKa = 6.11STRR19 pKa = 11.84EE20 pKa = 3.65YY21 pKa = 10.59NRR23 pKa = 11.84CRR25 pKa = 11.84ICGRR29 pKa = 11.84PHH31 pKa = 6.75AYY33 pKa = 9.43LRR35 pKa = 11.84KK36 pKa = 9.69FGICRR41 pKa = 11.84ICFRR45 pKa = 11.84EE46 pKa = 3.81LAYY49 pKa = 10.0KK50 pKa = 10.86GEE52 pKa = 4.16IPGVRR57 pKa = 11.84KK58 pKa = 10.13ASWW61 pKa = 2.78
MM1 pKa = 7.35ARR3 pKa = 11.84KK4 pKa = 9.59ALVVKK9 pKa = 7.44QQRR12 pKa = 11.84KK13 pKa = 6.5QKK15 pKa = 10.41YY16 pKa = 6.11STRR19 pKa = 11.84EE20 pKa = 3.65YY21 pKa = 10.59NRR23 pKa = 11.84CRR25 pKa = 11.84ICGRR29 pKa = 11.84PHH31 pKa = 6.75AYY33 pKa = 9.43LRR35 pKa = 11.84KK36 pKa = 9.69FGICRR41 pKa = 11.84ICFRR45 pKa = 11.84EE46 pKa = 3.81LAYY49 pKa = 10.0KK50 pKa = 10.86GEE52 pKa = 4.16IPGVRR57 pKa = 11.84KK58 pKa = 10.13ASWW61 pKa = 2.78
Molecular weight: 7.32 kDa
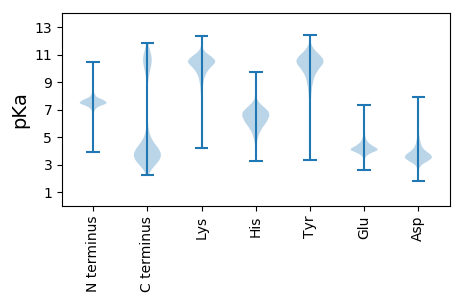
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1330799 |
26 |
2067 |
312.5 |
35.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.469 ± 0.043 | 1.121 ± 0.017 |
5.521 ± 0.03 | 7.715 ± 0.046 |
4.258 ± 0.031 | 6.724 ± 0.04 |
1.654 ± 0.016 | 9.914 ± 0.044 |
8.972 ± 0.039 | 8.961 ± 0.034 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.926 ± 0.016 | 5.859 ± 0.036 |
2.931 ± 0.023 | 2.205 ± 0.019 |
3.498 ± 0.028 | 5.931 ± 0.031 |
4.872 ± 0.026 | 6.708 ± 0.035 |
0.713 ± 0.011 | 4.047 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
