
Sanxia Water Strider Virus 2
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Phasmaviridae; Sawastrivirus; Sanxia sawastrivirus
Average proteome isoelectric point is 6.89
Get precalculated fractions of proteins
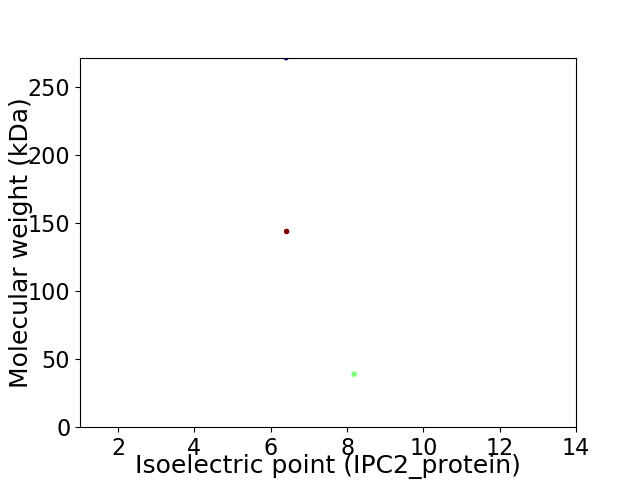
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
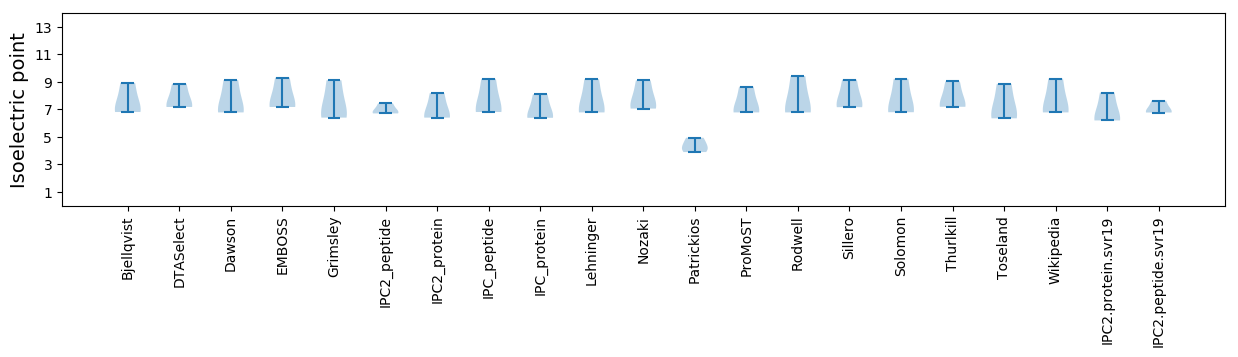
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L4AB05|A0A1L4AB05_9VIRU Nucleocapsid protein OS=Sanxia Water Strider Virus 2 OX=1608061 GN=N PE=4 SV=1
MM1 pKa = 7.53ALIIVLTLFAALSNAEE17 pKa = 4.26YY18 pKa = 10.7LRR20 pKa = 11.84EE21 pKa = 4.03TYY23 pKa = 10.3DD24 pKa = 2.67WYY26 pKa = 9.94TAGKK30 pKa = 9.7NNDD33 pKa = 2.7ISGLHH38 pKa = 6.05NLIQKK43 pKa = 8.51EE44 pKa = 4.24PKK46 pKa = 9.91NMEE49 pKa = 4.1IKK51 pKa = 10.5SEE53 pKa = 4.01SCEE56 pKa = 3.93NGGCRR61 pKa = 11.84LSLEE65 pKa = 4.32RR66 pKa = 11.84NFNILTSKK74 pKa = 10.76GFGINFYY81 pKa = 9.82VDD83 pKa = 2.85EE84 pKa = 4.46MGYY87 pKa = 10.58VNNTILVQVMDD98 pKa = 3.55TTFNVTAKK106 pKa = 10.56YY107 pKa = 10.47KK108 pKa = 9.62YY109 pKa = 6.19TTPACKK115 pKa = 10.12YY116 pKa = 8.68FYY118 pKa = 10.9RR119 pKa = 11.84NAFFNYY125 pKa = 9.09NQQKK129 pKa = 9.6YY130 pKa = 9.29ANCKK134 pKa = 9.97SGLGTDD140 pKa = 4.01DD141 pKa = 3.8FMSIAEE147 pKa = 4.06MQAYY151 pKa = 9.14GQFGVDD157 pKa = 3.36IEE159 pKa = 4.49VADD162 pKa = 5.07RR163 pKa = 11.84KK164 pKa = 8.79PTLLDD169 pKa = 3.5LGGWSEE175 pKa = 3.96EE176 pKa = 4.32AGLCGHH182 pKa = 7.35WWPTVLKK189 pKa = 9.48TCFEE193 pKa = 4.2KK194 pKa = 11.14ALICSLKK201 pKa = 10.46EE202 pKa = 4.05SFVLFALEE210 pKa = 3.92PEE212 pKa = 4.65IVFEE216 pKa = 3.45MWIRR220 pKa = 11.84VTVGQNTYY228 pKa = 11.17DD229 pKa = 3.36VVYY232 pKa = 10.37EE233 pKa = 4.22GNTNEE238 pKa = 3.71MRR240 pKa = 11.84TIEE243 pKa = 4.17VSEE246 pKa = 4.72GIYY249 pKa = 10.6LDD251 pKa = 3.79INPIGQLVNPINSKK265 pKa = 10.93AIGAKK270 pKa = 8.97IHH272 pKa = 5.69YY273 pKa = 7.31EE274 pKa = 3.95KK275 pKa = 11.03SNKK278 pKa = 6.77TAKK281 pKa = 10.66LEE283 pKa = 4.31SQWVYY288 pKa = 11.03LDD290 pKa = 4.31SYY292 pKa = 9.53PDD294 pKa = 3.15SGQIVSQGVGKK305 pKa = 9.57FQLVNLNNIKK315 pKa = 9.2STISNKK321 pKa = 10.02NNLNCDD327 pKa = 3.41GTIQSCTKK335 pKa = 10.0NLNRR339 pKa = 11.84KK340 pKa = 8.72LRR342 pKa = 11.84TKK344 pKa = 8.68QTNTGFSEE352 pKa = 4.27KK353 pKa = 10.1KK354 pKa = 10.49LLDD357 pKa = 3.29IYY359 pKa = 10.56SIKK362 pKa = 10.98NSDD365 pKa = 3.51TVEE368 pKa = 4.61DD369 pKa = 4.5DD370 pKa = 5.14LIIIAPNSSLQNLKK384 pKa = 11.01GGDD387 pKa = 3.59TCCIKK392 pKa = 10.7LISPSEE398 pKa = 4.04RR399 pKa = 11.84FLDD402 pKa = 4.08SIVNNYY408 pKa = 9.46EE409 pKa = 3.68EE410 pKa = 4.32ANKK413 pKa = 10.21VLMTKK418 pKa = 9.51MPILPSSFKK427 pKa = 10.4ISVSGTPEE435 pKa = 3.83LDD437 pKa = 3.19SFHH440 pKa = 7.02FKK442 pKa = 10.94NLLSVPILATLRR454 pKa = 11.84TKK456 pKa = 10.91DD457 pKa = 3.4LFLPFSNKK465 pKa = 8.44EE466 pKa = 3.82FEE468 pKa = 4.13ITNLIISSMTYY479 pKa = 10.06FEE481 pKa = 4.34NAAGSFFEE489 pKa = 4.3MSFDD493 pKa = 3.72YY494 pKa = 10.9SGPPGNIRR502 pKa = 11.84IDD504 pKa = 3.61SKK506 pKa = 11.34DD507 pKa = 3.21INILTKK513 pKa = 10.06EE514 pKa = 4.31LYY516 pKa = 9.72ISSSGKK522 pKa = 8.67YY523 pKa = 8.11TDD525 pKa = 3.39TVGFKK530 pKa = 10.72SKK532 pKa = 11.09GIMALKK538 pKa = 9.03PTICVGDD545 pKa = 3.66AAHH548 pKa = 6.47CVKK551 pKa = 9.84PAKK554 pKa = 10.17IINTKK559 pKa = 9.34TKK561 pKa = 10.31DD562 pKa = 3.61GSVDD566 pKa = 3.44DD567 pKa = 4.6PMTGEE572 pKa = 4.25EE573 pKa = 4.8DD574 pKa = 3.65SLINQITTMFKK585 pKa = 10.77LYY587 pKa = 9.95IANIWFCIFMTIIWLFDD604 pKa = 3.3IVLSAVLIIIIVKK617 pKa = 9.77LCGRR621 pKa = 11.84STLMLTVLCCSKK633 pKa = 10.34RR634 pKa = 11.84GEE636 pKa = 4.34SIEE639 pKa = 4.01TSKK642 pKa = 10.44ICEE645 pKa = 4.19LEE647 pKa = 3.98KK648 pKa = 11.12VGTLSSIIRR657 pKa = 11.84FDD659 pKa = 4.18HH660 pKa = 5.67IVGVKK665 pKa = 10.03LEE667 pKa = 4.1KK668 pKa = 11.13SMTDD672 pKa = 3.69LSWMINDD679 pKa = 3.42QMKK682 pKa = 10.35VLSINMPTKK691 pKa = 10.69SSLLEE696 pKa = 3.78PKK698 pKa = 10.07VKK700 pKa = 10.39NFYY703 pKa = 10.5HH704 pKa = 6.45VGVANYY710 pKa = 9.14SHH712 pKa = 7.43EE713 pKa = 4.85DD714 pKa = 3.47GAHH717 pKa = 6.41FLLPLYY723 pKa = 9.87GANPWEE729 pKa = 5.18IGTCDD734 pKa = 5.12DD735 pKa = 5.45DD736 pKa = 6.41NDD738 pKa = 4.7DD739 pKa = 4.04SGCNLEE745 pKa = 5.56EE746 pKa = 3.93ITADD750 pKa = 3.05TVTRR754 pKa = 11.84NTNFLLNLWNQVPRR768 pKa = 11.84VTQFLYY774 pKa = 10.27IKK776 pKa = 8.27STSYY780 pKa = 10.24PRR782 pKa = 11.84HH783 pKa = 6.33HH784 pKa = 7.26YY785 pKa = 11.08DD786 pKa = 3.67NGCSTYY792 pKa = 10.2TVALISASGTSSYY805 pKa = 10.56ATILAYY811 pKa = 9.42STAYY815 pKa = 9.53TGINLIEE822 pKa = 3.9VSKK825 pKa = 10.83FGNSKK830 pKa = 9.22IFAVEE835 pKa = 3.99VVAVASYY842 pKa = 11.0KK843 pKa = 10.37MNIDD847 pKa = 3.39VVKK850 pKa = 10.89NNVRR854 pKa = 11.84SMIKK858 pKa = 9.83ADD860 pKa = 3.83EE861 pKa = 4.23NVVISPINKK870 pKa = 9.17QSIDD874 pKa = 3.73AIGRR878 pKa = 11.84FYY880 pKa = 11.18FLITCPSGCVIYY892 pKa = 10.99NPLVNKK898 pKa = 10.1NGKK901 pKa = 10.04AEE903 pKa = 4.18MIKK906 pKa = 8.94ITDD909 pKa = 3.86SLLKK913 pKa = 9.76HH914 pKa = 6.13QIYY917 pKa = 10.81NDD919 pKa = 3.25VSVFRR924 pKa = 11.84SFWKK928 pKa = 8.96TNKK931 pKa = 9.59VNILSQEE938 pKa = 3.95NKK940 pKa = 9.53LSKK943 pKa = 10.64DD944 pKa = 2.95IVIRR948 pKa = 11.84YY949 pKa = 8.43QNDD952 pKa = 3.28YY953 pKa = 10.5NKK955 pKa = 10.57KK956 pKa = 10.31FGLDD960 pKa = 3.48DD961 pKa = 3.64VKK963 pKa = 11.47SDD965 pKa = 3.58VEE967 pKa = 4.41SFINNKK973 pKa = 7.09WYY975 pKa = 10.35RR976 pKa = 11.84YY977 pKa = 8.08NLQKK981 pKa = 10.66ISDD984 pKa = 4.06NTDD987 pKa = 3.03SNGDD991 pKa = 3.02ITLDD995 pKa = 4.19DD996 pKa = 4.15NQLHH1000 pKa = 5.36QLIDD1004 pKa = 3.63PTSDD1008 pKa = 4.33FIMTDD1013 pKa = 3.3DD1014 pKa = 5.52GILDD1018 pKa = 3.59VTLNGPISMNCHH1030 pKa = 4.66STGVGARR1037 pKa = 11.84CRR1039 pKa = 11.84HH1040 pKa = 4.86EE1041 pKa = 4.25TGNMIAHH1048 pKa = 7.31FYY1050 pKa = 8.01NTRR1053 pKa = 11.84TDD1055 pKa = 3.08INKK1058 pKa = 9.93FGSFKK1063 pKa = 11.26DD1064 pKa = 3.26NGQSYY1069 pKa = 10.07KK1070 pKa = 10.46YY1071 pKa = 9.48EE1072 pKa = 4.14KK1073 pKa = 10.18SDD1075 pKa = 3.63YY1076 pKa = 10.74KK1077 pKa = 11.27VGLLPRR1083 pKa = 11.84KK1084 pKa = 8.96EE1085 pKa = 3.95PCKK1088 pKa = 10.31GKK1090 pKa = 10.86GYY1092 pKa = 9.98IDD1094 pKa = 3.45SQSRR1098 pKa = 11.84FLCSHH1103 pKa = 7.13DD1104 pKa = 3.36CCMVSTNNWNNYY1116 pKa = 9.03FLVSANKK1123 pKa = 9.54PFVPNRR1129 pKa = 11.84TLEE1132 pKa = 4.52SINMWQSNKK1141 pKa = 8.38DD1142 pKa = 3.08TKK1144 pKa = 11.07LYY1146 pKa = 10.5FKK1148 pKa = 10.74SPYY1151 pKa = 10.31QCMTDD1156 pKa = 3.3TFSQTLDD1163 pKa = 3.3CALEE1167 pKa = 4.06KK1168 pKa = 10.62FSAEE1172 pKa = 4.05FYY1174 pKa = 9.77ISVFIVPMFLIIYY1187 pKa = 8.45FIMLICCACVKK1198 pKa = 10.36KK1199 pKa = 10.57SYY1201 pKa = 10.11RR1202 pKa = 11.84YY1203 pKa = 9.72KK1204 pKa = 10.95KK1205 pKa = 10.27LGNSAKK1211 pKa = 10.27KK1212 pKa = 10.39GRR1214 pKa = 11.84WLIADD1219 pKa = 3.56MADD1222 pKa = 3.32NIMKK1226 pKa = 9.93GKK1228 pKa = 8.85PKK1230 pKa = 10.32FWIRR1234 pKa = 11.84PDD1236 pKa = 3.77NSYY1239 pKa = 10.84EE1240 pKa = 3.81WDD1242 pKa = 4.08DD1243 pKa = 4.41NDD1245 pKa = 3.17IGPYY1249 pKa = 10.01KK1250 pKa = 10.01RR1251 pKa = 11.84TTTGRR1256 pKa = 11.84FRR1258 pKa = 11.84YY1259 pKa = 9.51NKK1261 pKa = 9.71LKK1263 pKa = 10.57AWSNEE1268 pKa = 3.13LDD1270 pKa = 3.29
MM1 pKa = 7.53ALIIVLTLFAALSNAEE17 pKa = 4.26YY18 pKa = 10.7LRR20 pKa = 11.84EE21 pKa = 4.03TYY23 pKa = 10.3DD24 pKa = 2.67WYY26 pKa = 9.94TAGKK30 pKa = 9.7NNDD33 pKa = 2.7ISGLHH38 pKa = 6.05NLIQKK43 pKa = 8.51EE44 pKa = 4.24PKK46 pKa = 9.91NMEE49 pKa = 4.1IKK51 pKa = 10.5SEE53 pKa = 4.01SCEE56 pKa = 3.93NGGCRR61 pKa = 11.84LSLEE65 pKa = 4.32RR66 pKa = 11.84NFNILTSKK74 pKa = 10.76GFGINFYY81 pKa = 9.82VDD83 pKa = 2.85EE84 pKa = 4.46MGYY87 pKa = 10.58VNNTILVQVMDD98 pKa = 3.55TTFNVTAKK106 pKa = 10.56YY107 pKa = 10.47KK108 pKa = 9.62YY109 pKa = 6.19TTPACKK115 pKa = 10.12YY116 pKa = 8.68FYY118 pKa = 10.9RR119 pKa = 11.84NAFFNYY125 pKa = 9.09NQQKK129 pKa = 9.6YY130 pKa = 9.29ANCKK134 pKa = 9.97SGLGTDD140 pKa = 4.01DD141 pKa = 3.8FMSIAEE147 pKa = 4.06MQAYY151 pKa = 9.14GQFGVDD157 pKa = 3.36IEE159 pKa = 4.49VADD162 pKa = 5.07RR163 pKa = 11.84KK164 pKa = 8.79PTLLDD169 pKa = 3.5LGGWSEE175 pKa = 3.96EE176 pKa = 4.32AGLCGHH182 pKa = 7.35WWPTVLKK189 pKa = 9.48TCFEE193 pKa = 4.2KK194 pKa = 11.14ALICSLKK201 pKa = 10.46EE202 pKa = 4.05SFVLFALEE210 pKa = 3.92PEE212 pKa = 4.65IVFEE216 pKa = 3.45MWIRR220 pKa = 11.84VTVGQNTYY228 pKa = 11.17DD229 pKa = 3.36VVYY232 pKa = 10.37EE233 pKa = 4.22GNTNEE238 pKa = 3.71MRR240 pKa = 11.84TIEE243 pKa = 4.17VSEE246 pKa = 4.72GIYY249 pKa = 10.6LDD251 pKa = 3.79INPIGQLVNPINSKK265 pKa = 10.93AIGAKK270 pKa = 8.97IHH272 pKa = 5.69YY273 pKa = 7.31EE274 pKa = 3.95KK275 pKa = 11.03SNKK278 pKa = 6.77TAKK281 pKa = 10.66LEE283 pKa = 4.31SQWVYY288 pKa = 11.03LDD290 pKa = 4.31SYY292 pKa = 9.53PDD294 pKa = 3.15SGQIVSQGVGKK305 pKa = 9.57FQLVNLNNIKK315 pKa = 9.2STISNKK321 pKa = 10.02NNLNCDD327 pKa = 3.41GTIQSCTKK335 pKa = 10.0NLNRR339 pKa = 11.84KK340 pKa = 8.72LRR342 pKa = 11.84TKK344 pKa = 8.68QTNTGFSEE352 pKa = 4.27KK353 pKa = 10.1KK354 pKa = 10.49LLDD357 pKa = 3.29IYY359 pKa = 10.56SIKK362 pKa = 10.98NSDD365 pKa = 3.51TVEE368 pKa = 4.61DD369 pKa = 4.5DD370 pKa = 5.14LIIIAPNSSLQNLKK384 pKa = 11.01GGDD387 pKa = 3.59TCCIKK392 pKa = 10.7LISPSEE398 pKa = 4.04RR399 pKa = 11.84FLDD402 pKa = 4.08SIVNNYY408 pKa = 9.46EE409 pKa = 3.68EE410 pKa = 4.32ANKK413 pKa = 10.21VLMTKK418 pKa = 9.51MPILPSSFKK427 pKa = 10.4ISVSGTPEE435 pKa = 3.83LDD437 pKa = 3.19SFHH440 pKa = 7.02FKK442 pKa = 10.94NLLSVPILATLRR454 pKa = 11.84TKK456 pKa = 10.91DD457 pKa = 3.4LFLPFSNKK465 pKa = 8.44EE466 pKa = 3.82FEE468 pKa = 4.13ITNLIISSMTYY479 pKa = 10.06FEE481 pKa = 4.34NAAGSFFEE489 pKa = 4.3MSFDD493 pKa = 3.72YY494 pKa = 10.9SGPPGNIRR502 pKa = 11.84IDD504 pKa = 3.61SKK506 pKa = 11.34DD507 pKa = 3.21INILTKK513 pKa = 10.06EE514 pKa = 4.31LYY516 pKa = 9.72ISSSGKK522 pKa = 8.67YY523 pKa = 8.11TDD525 pKa = 3.39TVGFKK530 pKa = 10.72SKK532 pKa = 11.09GIMALKK538 pKa = 9.03PTICVGDD545 pKa = 3.66AAHH548 pKa = 6.47CVKK551 pKa = 9.84PAKK554 pKa = 10.17IINTKK559 pKa = 9.34TKK561 pKa = 10.31DD562 pKa = 3.61GSVDD566 pKa = 3.44DD567 pKa = 4.6PMTGEE572 pKa = 4.25EE573 pKa = 4.8DD574 pKa = 3.65SLINQITTMFKK585 pKa = 10.77LYY587 pKa = 9.95IANIWFCIFMTIIWLFDD604 pKa = 3.3IVLSAVLIIIIVKK617 pKa = 9.77LCGRR621 pKa = 11.84STLMLTVLCCSKK633 pKa = 10.34RR634 pKa = 11.84GEE636 pKa = 4.34SIEE639 pKa = 4.01TSKK642 pKa = 10.44ICEE645 pKa = 4.19LEE647 pKa = 3.98KK648 pKa = 11.12VGTLSSIIRR657 pKa = 11.84FDD659 pKa = 4.18HH660 pKa = 5.67IVGVKK665 pKa = 10.03LEE667 pKa = 4.1KK668 pKa = 11.13SMTDD672 pKa = 3.69LSWMINDD679 pKa = 3.42QMKK682 pKa = 10.35VLSINMPTKK691 pKa = 10.69SSLLEE696 pKa = 3.78PKK698 pKa = 10.07VKK700 pKa = 10.39NFYY703 pKa = 10.5HH704 pKa = 6.45VGVANYY710 pKa = 9.14SHH712 pKa = 7.43EE713 pKa = 4.85DD714 pKa = 3.47GAHH717 pKa = 6.41FLLPLYY723 pKa = 9.87GANPWEE729 pKa = 5.18IGTCDD734 pKa = 5.12DD735 pKa = 5.45DD736 pKa = 6.41NDD738 pKa = 4.7DD739 pKa = 4.04SGCNLEE745 pKa = 5.56EE746 pKa = 3.93ITADD750 pKa = 3.05TVTRR754 pKa = 11.84NTNFLLNLWNQVPRR768 pKa = 11.84VTQFLYY774 pKa = 10.27IKK776 pKa = 8.27STSYY780 pKa = 10.24PRR782 pKa = 11.84HH783 pKa = 6.33HH784 pKa = 7.26YY785 pKa = 11.08DD786 pKa = 3.67NGCSTYY792 pKa = 10.2TVALISASGTSSYY805 pKa = 10.56ATILAYY811 pKa = 9.42STAYY815 pKa = 9.53TGINLIEE822 pKa = 3.9VSKK825 pKa = 10.83FGNSKK830 pKa = 9.22IFAVEE835 pKa = 3.99VVAVASYY842 pKa = 11.0KK843 pKa = 10.37MNIDD847 pKa = 3.39VVKK850 pKa = 10.89NNVRR854 pKa = 11.84SMIKK858 pKa = 9.83ADD860 pKa = 3.83EE861 pKa = 4.23NVVISPINKK870 pKa = 9.17QSIDD874 pKa = 3.73AIGRR878 pKa = 11.84FYY880 pKa = 11.18FLITCPSGCVIYY892 pKa = 10.99NPLVNKK898 pKa = 10.1NGKK901 pKa = 10.04AEE903 pKa = 4.18MIKK906 pKa = 8.94ITDD909 pKa = 3.86SLLKK913 pKa = 9.76HH914 pKa = 6.13QIYY917 pKa = 10.81NDD919 pKa = 3.25VSVFRR924 pKa = 11.84SFWKK928 pKa = 8.96TNKK931 pKa = 9.59VNILSQEE938 pKa = 3.95NKK940 pKa = 9.53LSKK943 pKa = 10.64DD944 pKa = 2.95IVIRR948 pKa = 11.84YY949 pKa = 8.43QNDD952 pKa = 3.28YY953 pKa = 10.5NKK955 pKa = 10.57KK956 pKa = 10.31FGLDD960 pKa = 3.48DD961 pKa = 3.64VKK963 pKa = 11.47SDD965 pKa = 3.58VEE967 pKa = 4.41SFINNKK973 pKa = 7.09WYY975 pKa = 10.35RR976 pKa = 11.84YY977 pKa = 8.08NLQKK981 pKa = 10.66ISDD984 pKa = 4.06NTDD987 pKa = 3.03SNGDD991 pKa = 3.02ITLDD995 pKa = 4.19DD996 pKa = 4.15NQLHH1000 pKa = 5.36QLIDD1004 pKa = 3.63PTSDD1008 pKa = 4.33FIMTDD1013 pKa = 3.3DD1014 pKa = 5.52GILDD1018 pKa = 3.59VTLNGPISMNCHH1030 pKa = 4.66STGVGARR1037 pKa = 11.84CRR1039 pKa = 11.84HH1040 pKa = 4.86EE1041 pKa = 4.25TGNMIAHH1048 pKa = 7.31FYY1050 pKa = 8.01NTRR1053 pKa = 11.84TDD1055 pKa = 3.08INKK1058 pKa = 9.93FGSFKK1063 pKa = 11.26DD1064 pKa = 3.26NGQSYY1069 pKa = 10.07KK1070 pKa = 10.46YY1071 pKa = 9.48EE1072 pKa = 4.14KK1073 pKa = 10.18SDD1075 pKa = 3.63YY1076 pKa = 10.74KK1077 pKa = 11.27VGLLPRR1083 pKa = 11.84KK1084 pKa = 8.96EE1085 pKa = 3.95PCKK1088 pKa = 10.31GKK1090 pKa = 10.86GYY1092 pKa = 9.98IDD1094 pKa = 3.45SQSRR1098 pKa = 11.84FLCSHH1103 pKa = 7.13DD1104 pKa = 3.36CCMVSTNNWNNYY1116 pKa = 9.03FLVSANKK1123 pKa = 9.54PFVPNRR1129 pKa = 11.84TLEE1132 pKa = 4.52SINMWQSNKK1141 pKa = 8.38DD1142 pKa = 3.08TKK1144 pKa = 11.07LYY1146 pKa = 10.5FKK1148 pKa = 10.74SPYY1151 pKa = 10.31QCMTDD1156 pKa = 3.3TFSQTLDD1163 pKa = 3.3CALEE1167 pKa = 4.06KK1168 pKa = 10.62FSAEE1172 pKa = 4.05FYY1174 pKa = 9.77ISVFIVPMFLIIYY1187 pKa = 8.45FIMLICCACVKK1198 pKa = 10.36KK1199 pKa = 10.57SYY1201 pKa = 10.11RR1202 pKa = 11.84YY1203 pKa = 9.72KK1204 pKa = 10.95KK1205 pKa = 10.27LGNSAKK1211 pKa = 10.27KK1212 pKa = 10.39GRR1214 pKa = 11.84WLIADD1219 pKa = 3.56MADD1222 pKa = 3.32NIMKK1226 pKa = 9.93GKK1228 pKa = 8.85PKK1230 pKa = 10.32FWIRR1234 pKa = 11.84PDD1236 pKa = 3.77NSYY1239 pKa = 10.84EE1240 pKa = 3.81WDD1242 pKa = 4.08DD1243 pKa = 4.41NDD1245 pKa = 3.17IGPYY1249 pKa = 10.01KK1250 pKa = 10.01RR1251 pKa = 11.84TTTGRR1256 pKa = 11.84FRR1258 pKa = 11.84YY1259 pKa = 9.51NKK1261 pKa = 9.71LKK1263 pKa = 10.57AWSNEE1268 pKa = 3.13LDD1270 pKa = 3.29
Molecular weight: 143.84 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L4AB05|A0A1L4AB05_9VIRU Nucleocapsid protein OS=Sanxia Water Strider Virus 2 OX=1608061 GN=N PE=4 SV=1
MM1 pKa = 7.07STTIKK6 pKa = 10.49SEE8 pKa = 3.7QFTNVQQVVIPDD20 pKa = 3.26KK21 pKa = 10.61WADD24 pKa = 3.48KK25 pKa = 10.29LASEE29 pKa = 4.99SEE31 pKa = 4.34VTYY34 pKa = 9.74STNKK38 pKa = 10.11LEE40 pKa = 4.23VYY42 pKa = 9.82QVIFKK47 pKa = 11.03KK48 pKa = 10.75LMTRR52 pKa = 11.84VPVIGSDD59 pKa = 3.53MEE61 pKa = 4.33VDD63 pKa = 4.45LDD65 pKa = 4.14TPPAAYY71 pKa = 10.15AFATYY76 pKa = 8.46VTSDD80 pKa = 3.43FYY82 pKa = 11.32RR83 pKa = 11.84DD84 pKa = 3.55QVSKK88 pKa = 10.08STGEE92 pKa = 3.77VKK94 pKa = 10.84YY95 pKa = 10.46LLKK98 pKa = 10.74VGQQEE103 pKa = 3.95IPVYY107 pKa = 8.11STRR110 pKa = 11.84KK111 pKa = 9.22SAQKK115 pKa = 10.69LDD117 pKa = 4.13DD118 pKa = 4.13GLISVDD124 pKa = 3.98DD125 pKa = 3.91ATLASIMLQNTVVKK139 pKa = 10.19QFKK142 pKa = 9.03EE143 pKa = 4.39KK144 pKa = 10.28KK145 pKa = 9.03SAWAMTPLSRR155 pKa = 11.84ISMTNEE161 pKa = 3.94GIEE164 pKa = 4.02ALADD168 pKa = 3.53EE169 pKa = 5.16MKK171 pKa = 10.97VSDD174 pKa = 3.63TTIVKK179 pKa = 8.59MLNMSTARR187 pKa = 11.84KK188 pKa = 9.69SYY190 pKa = 9.63QCSDD194 pKa = 3.76IIGSGYY200 pKa = 10.75SSDD203 pKa = 3.65FAVASILRR211 pKa = 11.84SNFLNDD217 pKa = 3.42RR218 pKa = 11.84NKK220 pKa = 9.19VTMLQKK226 pKa = 10.59QISDD230 pKa = 3.67KK231 pKa = 11.27NIARR235 pKa = 11.84CVRR238 pKa = 11.84AAPIDD243 pKa = 3.77KK244 pKa = 10.66DD245 pKa = 3.6RR246 pKa = 11.84VAMILSCMSGISNDD260 pKa = 3.17VDD262 pKa = 3.59KK263 pKa = 11.67EE264 pKa = 4.15EE265 pKa = 5.0ILMKK269 pKa = 10.6VKK271 pKa = 10.21NLQSIGVKK279 pKa = 9.86RR280 pKa = 11.84RR281 pKa = 11.84AGLFEE286 pKa = 4.38EE287 pKa = 4.51HH288 pKa = 6.08TKK290 pKa = 10.53AHH292 pKa = 5.69SVEE295 pKa = 4.17NFPTEE300 pKa = 4.27AEE302 pKa = 4.12TLSYY306 pKa = 11.21LEE308 pKa = 4.38ARR310 pKa = 11.84GIQDD314 pKa = 3.23RR315 pKa = 11.84QNLKK319 pKa = 10.73KK320 pKa = 10.01IVKK323 pKa = 8.93EE324 pKa = 4.08VKK326 pKa = 10.01LNKK329 pKa = 10.18ALTKK333 pKa = 9.26STKK336 pKa = 8.37EE337 pKa = 3.64QILNKK342 pKa = 9.8IAEE345 pKa = 4.33KK346 pKa = 10.6LAA348 pKa = 3.9
MM1 pKa = 7.07STTIKK6 pKa = 10.49SEE8 pKa = 3.7QFTNVQQVVIPDD20 pKa = 3.26KK21 pKa = 10.61WADD24 pKa = 3.48KK25 pKa = 10.29LASEE29 pKa = 4.99SEE31 pKa = 4.34VTYY34 pKa = 9.74STNKK38 pKa = 10.11LEE40 pKa = 4.23VYY42 pKa = 9.82QVIFKK47 pKa = 11.03KK48 pKa = 10.75LMTRR52 pKa = 11.84VPVIGSDD59 pKa = 3.53MEE61 pKa = 4.33VDD63 pKa = 4.45LDD65 pKa = 4.14TPPAAYY71 pKa = 10.15AFATYY76 pKa = 8.46VTSDD80 pKa = 3.43FYY82 pKa = 11.32RR83 pKa = 11.84DD84 pKa = 3.55QVSKK88 pKa = 10.08STGEE92 pKa = 3.77VKK94 pKa = 10.84YY95 pKa = 10.46LLKK98 pKa = 10.74VGQQEE103 pKa = 3.95IPVYY107 pKa = 8.11STRR110 pKa = 11.84KK111 pKa = 9.22SAQKK115 pKa = 10.69LDD117 pKa = 4.13DD118 pKa = 4.13GLISVDD124 pKa = 3.98DD125 pKa = 3.91ATLASIMLQNTVVKK139 pKa = 10.19QFKK142 pKa = 9.03EE143 pKa = 4.39KK144 pKa = 10.28KK145 pKa = 9.03SAWAMTPLSRR155 pKa = 11.84ISMTNEE161 pKa = 3.94GIEE164 pKa = 4.02ALADD168 pKa = 3.53EE169 pKa = 5.16MKK171 pKa = 10.97VSDD174 pKa = 3.63TTIVKK179 pKa = 8.59MLNMSTARR187 pKa = 11.84KK188 pKa = 9.69SYY190 pKa = 9.63QCSDD194 pKa = 3.76IIGSGYY200 pKa = 10.75SSDD203 pKa = 3.65FAVASILRR211 pKa = 11.84SNFLNDD217 pKa = 3.42RR218 pKa = 11.84NKK220 pKa = 9.19VTMLQKK226 pKa = 10.59QISDD230 pKa = 3.67KK231 pKa = 11.27NIARR235 pKa = 11.84CVRR238 pKa = 11.84AAPIDD243 pKa = 3.77KK244 pKa = 10.66DD245 pKa = 3.6RR246 pKa = 11.84VAMILSCMSGISNDD260 pKa = 3.17VDD262 pKa = 3.59KK263 pKa = 11.67EE264 pKa = 4.15EE265 pKa = 5.0ILMKK269 pKa = 10.6VKK271 pKa = 10.21NLQSIGVKK279 pKa = 9.86RR280 pKa = 11.84RR281 pKa = 11.84AGLFEE286 pKa = 4.38EE287 pKa = 4.51HH288 pKa = 6.08TKK290 pKa = 10.53AHH292 pKa = 5.69SVEE295 pKa = 4.17NFPTEE300 pKa = 4.27AEE302 pKa = 4.12TLSYY306 pKa = 11.21LEE308 pKa = 4.38ARR310 pKa = 11.84GIQDD314 pKa = 3.23RR315 pKa = 11.84QNLKK319 pKa = 10.73KK320 pKa = 10.01IVKK323 pKa = 8.93EE324 pKa = 4.08VKK326 pKa = 10.01LNKK329 pKa = 10.18ALTKK333 pKa = 9.26STKK336 pKa = 8.37EE337 pKa = 3.64QILNKK342 pKa = 9.8IAEE345 pKa = 4.33KK346 pKa = 10.6LAA348 pKa = 3.9
Molecular weight: 38.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3962 |
348 |
2344 |
1320.7 |
151.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.619 ± 0.586 | 1.489 ± 0.513 |
6.663 ± 0.269 | 6.032 ± 0.58 |
4.417 ± 0.345 | 4.619 ± 0.411 |
1.212 ± 0.119 | 8.253 ± 0.259 |
8.657 ± 0.358 | 8.758 ± 0.377 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.877 ± 0.163 | 7.774 ± 0.656 |
2.524 ± 0.273 | 3.231 ± 0.395 |
4.064 ± 0.6 | 7.496 ± 0.671 |
5.906 ± 0.4 | 5.704 ± 0.516 |
0.883 ± 0.265 | 4.821 ± 0.44 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
