
Slackia heliotrinireducens (strain ATCC 29202 / DSM 20476 / NCTC 11029 / RHS 1) (Peptococcus heliotrinreducens)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Coriobacteriia; Eggerthellales; Eggerthellaceae; Slackia; Slackia heliotrinireducens
Average proteome isoelectric point is 5.51
Get precalculated fractions of proteins
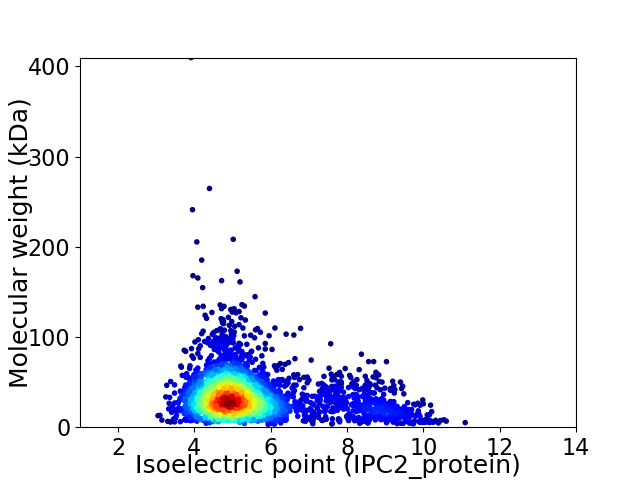
Virtual 2D-PAGE plot for 2750 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C7N1K1|C7N1K1_SLAHD tRNA N6-adenosine threonylcarbamoyltransferase OS=Slackia heliotrinireducens (strain ATCC 29202 / DSM 20476 / NCTC 11029 / RHS 1) OX=471855 GN=tsaD PE=3 SV=1
MM1 pKa = 7.59FCPNCGTQLPDD12 pKa = 3.44GSAFCGNCGNRR23 pKa = 11.84LNAAPNPAPAPGYY36 pKa = 9.84PPVAQPAGSFDD47 pKa = 3.14IKK49 pKa = 10.74KK50 pKa = 9.56FLPIIIGVAAAVLLLALFLIIRR72 pKa = 11.84PFGGGLSGEE81 pKa = 4.25NVGKK85 pKa = 10.42IMTMEE90 pKa = 4.18PEE92 pKa = 4.77KK93 pKa = 10.48IAKK96 pKa = 9.58QLDD99 pKa = 3.36KK100 pKa = 10.81MEE102 pKa = 4.32SVKK105 pKa = 10.43VDD107 pKa = 3.29GQRR110 pKa = 11.84LYY112 pKa = 11.23VSSEE116 pKa = 3.84EE117 pKa = 4.05FGDD120 pKa = 4.32LIKK123 pKa = 11.17DD124 pKa = 3.56ADD126 pKa = 4.32DD127 pKa = 4.22GLSKK131 pKa = 10.87KK132 pKa = 9.37EE133 pKa = 3.46AAAADD138 pKa = 4.31GEE140 pKa = 4.55WAFAVNPDD148 pKa = 4.2ARR150 pKa = 11.84DD151 pKa = 3.67LEE153 pKa = 5.85DD154 pKa = 5.44GDD156 pKa = 5.23DD157 pKa = 4.46LGSVSCFRR165 pKa = 11.84VYY167 pKa = 11.47ADD169 pKa = 4.61TDD171 pKa = 3.65DD172 pKa = 4.86LNCNKK177 pKa = 9.82IVDD180 pKa = 3.99YY181 pKa = 10.92AKK183 pKa = 11.31DD184 pKa = 3.21MGLDD188 pKa = 3.17VGTYY192 pKa = 9.49ACYY195 pKa = 10.62VGDD198 pKa = 4.07GGDD201 pKa = 3.54YY202 pKa = 11.18GYY204 pKa = 9.21LTAYY208 pKa = 10.21NDD210 pKa = 4.15DD211 pKa = 3.58YY212 pKa = 11.59SINLNVSIDD221 pKa = 3.44SHH223 pKa = 5.89ATVASATVYY232 pKa = 10.65TDD234 pKa = 3.96DD235 pKa = 6.29GYY237 pKa = 11.43DD238 pKa = 3.44LKK240 pKa = 11.15DD241 pKa = 3.7AQDD244 pKa = 3.52EE245 pKa = 4.7VEE247 pKa = 4.87NYY249 pKa = 10.86ADD251 pKa = 3.75NLEE254 pKa = 4.54DD255 pKa = 4.61EE256 pKa = 4.65MDD258 pKa = 3.51EE259 pKa = 4.1FDD261 pKa = 4.54YY262 pKa = 11.73EE263 pKa = 4.29EE264 pKa = 4.08IQTNVKK270 pKa = 10.15
MM1 pKa = 7.59FCPNCGTQLPDD12 pKa = 3.44GSAFCGNCGNRR23 pKa = 11.84LNAAPNPAPAPGYY36 pKa = 9.84PPVAQPAGSFDD47 pKa = 3.14IKK49 pKa = 10.74KK50 pKa = 9.56FLPIIIGVAAAVLLLALFLIIRR72 pKa = 11.84PFGGGLSGEE81 pKa = 4.25NVGKK85 pKa = 10.42IMTMEE90 pKa = 4.18PEE92 pKa = 4.77KK93 pKa = 10.48IAKK96 pKa = 9.58QLDD99 pKa = 3.36KK100 pKa = 10.81MEE102 pKa = 4.32SVKK105 pKa = 10.43VDD107 pKa = 3.29GQRR110 pKa = 11.84LYY112 pKa = 11.23VSSEE116 pKa = 3.84EE117 pKa = 4.05FGDD120 pKa = 4.32LIKK123 pKa = 11.17DD124 pKa = 3.56ADD126 pKa = 4.32DD127 pKa = 4.22GLSKK131 pKa = 10.87KK132 pKa = 9.37EE133 pKa = 3.46AAAADD138 pKa = 4.31GEE140 pKa = 4.55WAFAVNPDD148 pKa = 4.2ARR150 pKa = 11.84DD151 pKa = 3.67LEE153 pKa = 5.85DD154 pKa = 5.44GDD156 pKa = 5.23DD157 pKa = 4.46LGSVSCFRR165 pKa = 11.84VYY167 pKa = 11.47ADD169 pKa = 4.61TDD171 pKa = 3.65DD172 pKa = 4.86LNCNKK177 pKa = 9.82IVDD180 pKa = 3.99YY181 pKa = 10.92AKK183 pKa = 11.31DD184 pKa = 3.21MGLDD188 pKa = 3.17VGTYY192 pKa = 9.49ACYY195 pKa = 10.62VGDD198 pKa = 4.07GGDD201 pKa = 3.54YY202 pKa = 11.18GYY204 pKa = 9.21LTAYY208 pKa = 10.21NDD210 pKa = 4.15DD211 pKa = 3.58YY212 pKa = 11.59SINLNVSIDD221 pKa = 3.44SHH223 pKa = 5.89ATVASATVYY232 pKa = 10.65TDD234 pKa = 3.96DD235 pKa = 6.29GYY237 pKa = 11.43DD238 pKa = 3.44LKK240 pKa = 11.15DD241 pKa = 3.7AQDD244 pKa = 3.52EE245 pKa = 4.7VEE247 pKa = 4.87NYY249 pKa = 10.86ADD251 pKa = 3.75NLEE254 pKa = 4.54DD255 pKa = 4.61EE256 pKa = 4.65MDD258 pKa = 3.51EE259 pKa = 4.1FDD261 pKa = 4.54YY262 pKa = 11.73EE263 pKa = 4.29EE264 pKa = 4.08IQTNVKK270 pKa = 10.15
Molecular weight: 29.07 kDa
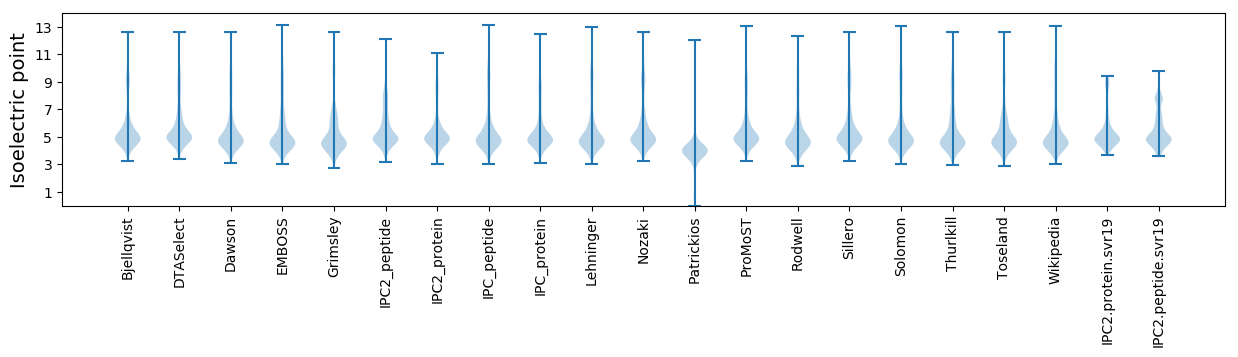
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C7N4A7|C7N4A7_SLAHD DMSO reductase iron-sulfur subunit OS=Slackia heliotrinireducens (strain ATCC 29202 / DSM 20476 / NCTC 11029 / RHS 1) OX=471855 GN=Shel_06830 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.42RR3 pKa = 11.84TYY5 pKa = 10.31QPNKK9 pKa = 7.52RR10 pKa = 11.84HH11 pKa = 5.83RR12 pKa = 11.84AKK14 pKa = 9.65THH16 pKa = 5.05GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.27GGRR28 pKa = 11.84AVLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.76GRR39 pKa = 11.84KK40 pKa = 8.98RR41 pKa = 11.84LTVV44 pKa = 3.11
MM1 pKa = 7.35KK2 pKa = 9.42RR3 pKa = 11.84TYY5 pKa = 10.31QPNKK9 pKa = 7.52RR10 pKa = 11.84HH11 pKa = 5.83RR12 pKa = 11.84AKK14 pKa = 9.65THH16 pKa = 5.05GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.27GGRR28 pKa = 11.84AVLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.76GRR39 pKa = 11.84KK40 pKa = 8.98RR41 pKa = 11.84LTVV44 pKa = 3.11
Molecular weight: 5.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
898832 |
31 |
3920 |
326.8 |
35.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.798 ± 0.067 | 1.65 ± 0.024 |
6.522 ± 0.04 | 6.882 ± 0.048 |
3.822 ± 0.026 | 7.985 ± 0.041 |
1.862 ± 0.022 | 5.308 ± 0.044 |
4.046 ± 0.034 | 8.689 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.937 ± 0.023 | 3.268 ± 0.032 |
4.224 ± 0.028 | 3.025 ± 0.027 |
5.411 ± 0.058 | 5.691 ± 0.036 |
5.535 ± 0.076 | 7.908 ± 0.043 |
1.173 ± 0.022 | 3.263 ± 0.036 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
