
Polyomavirus sp.
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; unclassified Polyomaviridae
Average proteome isoelectric point is 5.63
Get precalculated fractions of proteins
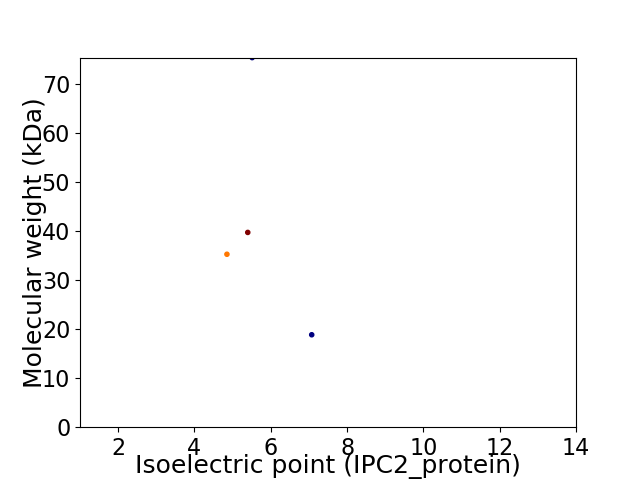
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1D8QQF4|A0A1D8QQF4_9POLY Minor capsid protein OS=Polyomavirus sp. OX=36362 PE=3 SV=1
MM1 pKa = 7.43FLLLGMGALLAVFAEE16 pKa = 4.8VIDD19 pKa = 4.21LAAATGLGIDD29 pKa = 5.06AILSAEE35 pKa = 4.64AITTSEE41 pKa = 4.1ALEE44 pKa = 3.88ALITNLVTLEE54 pKa = 4.19GFSEE58 pKa = 4.61AEE60 pKa = 3.94ALTAAGLTAQASAAFAEE77 pKa = 4.77LGSLYY82 pKa = 10.03PQILYY87 pKa = 10.94ALAFDD92 pKa = 4.26SLTVGTAVAAALAPNSFDD110 pKa = 3.7HH111 pKa = 6.35PTPIANLNMALVRR124 pKa = 11.84WVPDD128 pKa = 3.46FDD130 pKa = 4.67FLFPGFRR137 pKa = 11.84HH138 pKa = 6.31LARR141 pKa = 11.84FINYY145 pKa = 9.0IDD147 pKa = 3.73PTNWAPDD154 pKa = 3.44LYY156 pKa = 9.09EE157 pKa = 3.89TLGRR161 pKa = 11.84YY162 pKa = 7.29VWEE165 pKa = 4.31GLQRR169 pKa = 11.84RR170 pKa = 11.84GEE172 pKa = 4.08RR173 pKa = 11.84FIDD176 pKa = 3.68EE177 pKa = 4.02QIGNIEE183 pKa = 4.11HH184 pKa = 6.57VGRR187 pKa = 11.84EE188 pKa = 3.72AAIRR192 pKa = 11.84TVEE195 pKa = 4.17TFTEE199 pKa = 3.83ALARR203 pKa = 11.84YY204 pKa = 9.03FEE206 pKa = 4.13NARR209 pKa = 11.84WAVSYY214 pKa = 10.18IPHH217 pKa = 7.11HH218 pKa = 6.8IYY220 pKa = 10.65SGLEE224 pKa = 3.56KK225 pKa = 10.48YY226 pKa = 7.88YY227 pKa = 10.14TQLPPLNPIQIRR239 pKa = 11.84DD240 pKa = 3.4VSRR243 pKa = 11.84RR244 pKa = 11.84TGLPIPDD251 pKa = 3.35RR252 pKa = 11.84TVGPNGEE259 pKa = 4.34NVKK262 pKa = 10.17SAQYY266 pKa = 10.21VEE268 pKa = 5.1KK269 pKa = 10.43PEE271 pKa = 4.33SPGGAMQRR279 pKa = 11.84STPDD283 pKa = 2.44WMLPLILGLYY293 pKa = 10.19GDD295 pKa = 5.07VSPSWGEE302 pKa = 3.75TLQDD306 pKa = 4.88LEE308 pKa = 4.54NQDD311 pKa = 3.91GPLKK315 pKa = 10.35KK316 pKa = 9.93RR317 pKa = 11.84KK318 pKa = 8.28KK319 pKa = 7.7THH321 pKa = 5.25
MM1 pKa = 7.43FLLLGMGALLAVFAEE16 pKa = 4.8VIDD19 pKa = 4.21LAAATGLGIDD29 pKa = 5.06AILSAEE35 pKa = 4.64AITTSEE41 pKa = 4.1ALEE44 pKa = 3.88ALITNLVTLEE54 pKa = 4.19GFSEE58 pKa = 4.61AEE60 pKa = 3.94ALTAAGLTAQASAAFAEE77 pKa = 4.77LGSLYY82 pKa = 10.03PQILYY87 pKa = 10.94ALAFDD92 pKa = 4.26SLTVGTAVAAALAPNSFDD110 pKa = 3.7HH111 pKa = 6.35PTPIANLNMALVRR124 pKa = 11.84WVPDD128 pKa = 3.46FDD130 pKa = 4.67FLFPGFRR137 pKa = 11.84HH138 pKa = 6.31LARR141 pKa = 11.84FINYY145 pKa = 9.0IDD147 pKa = 3.73PTNWAPDD154 pKa = 3.44LYY156 pKa = 9.09EE157 pKa = 3.89TLGRR161 pKa = 11.84YY162 pKa = 7.29VWEE165 pKa = 4.31GLQRR169 pKa = 11.84RR170 pKa = 11.84GEE172 pKa = 4.08RR173 pKa = 11.84FIDD176 pKa = 3.68EE177 pKa = 4.02QIGNIEE183 pKa = 4.11HH184 pKa = 6.57VGRR187 pKa = 11.84EE188 pKa = 3.72AAIRR192 pKa = 11.84TVEE195 pKa = 4.17TFTEE199 pKa = 3.83ALARR203 pKa = 11.84YY204 pKa = 9.03FEE206 pKa = 4.13NARR209 pKa = 11.84WAVSYY214 pKa = 10.18IPHH217 pKa = 7.11HH218 pKa = 6.8IYY220 pKa = 10.65SGLEE224 pKa = 3.56KK225 pKa = 10.48YY226 pKa = 7.88YY227 pKa = 10.14TQLPPLNPIQIRR239 pKa = 11.84DD240 pKa = 3.4VSRR243 pKa = 11.84RR244 pKa = 11.84TGLPIPDD251 pKa = 3.35RR252 pKa = 11.84TVGPNGEE259 pKa = 4.34NVKK262 pKa = 10.17SAQYY266 pKa = 10.21VEE268 pKa = 5.1KK269 pKa = 10.43PEE271 pKa = 4.33SPGGAMQRR279 pKa = 11.84STPDD283 pKa = 2.44WMLPLILGLYY293 pKa = 10.19GDD295 pKa = 5.07VSPSWGEE302 pKa = 3.75TLQDD306 pKa = 4.88LEE308 pKa = 4.54NQDD311 pKa = 3.91GPLKK315 pKa = 10.35KK316 pKa = 9.93RR317 pKa = 11.84KK318 pKa = 8.28KK319 pKa = 7.7THH321 pKa = 5.25
Molecular weight: 35.29 kDa
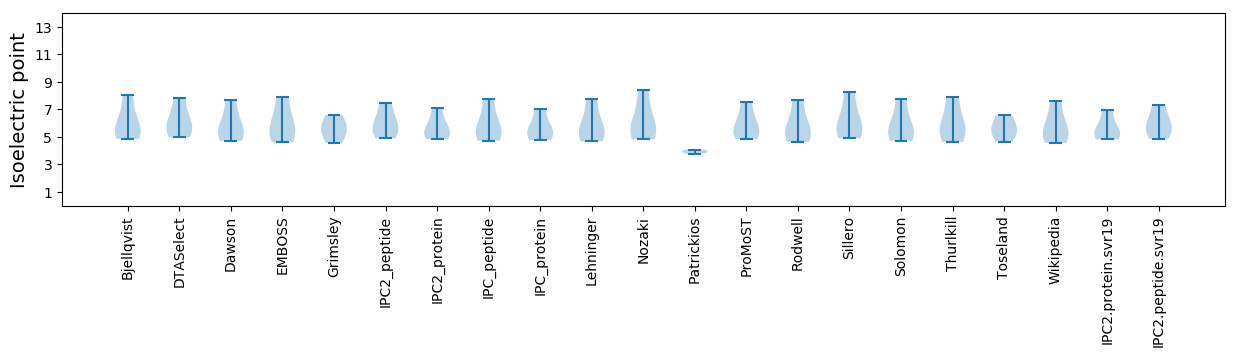
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1D8QQF4|A0A1D8QQF4_9POLY Minor capsid protein OS=Polyomavirus sp. OX=36362 PE=3 SV=1
MM1 pKa = 7.66DD2 pKa = 4.87HH3 pKa = 7.31ALTRR7 pKa = 11.84EE8 pKa = 4.04EE9 pKa = 4.51SVKK12 pKa = 10.73LMQLLKK18 pKa = 10.99LPMEE22 pKa = 4.41QYY24 pKa = 11.84GNFPLMRR31 pKa = 11.84RR32 pKa = 11.84AFLEE36 pKa = 3.98QCKK39 pKa = 10.09VLHH42 pKa = 6.76PDD44 pKa = 2.8KK45 pKa = 11.29GGNAEE50 pKa = 4.2LAKK53 pKa = 10.44EE54 pKa = 4.26LISLYY59 pKa = 10.42RR60 pKa = 11.84KK61 pKa = 10.03LEE63 pKa = 3.91AVIPPLNPNDD73 pKa = 3.78SFTTEE78 pKa = 3.77QVGVSDD84 pKa = 3.96SFVFYY89 pKa = 11.01LRR91 pKa = 11.84DD92 pKa = 3.14WSSCLKK98 pKa = 9.28GTEE101 pKa = 3.73VCICLFCLLRR111 pKa = 11.84DD112 pKa = 3.36SHH114 pKa = 6.52RR115 pKa = 11.84RR116 pKa = 11.84RR117 pKa = 11.84GSKK120 pKa = 8.99PKK122 pKa = 10.03CWGSCFCFEE131 pKa = 5.91CYY133 pKa = 9.67IAWFGLEE140 pKa = 3.75YY141 pKa = 10.01TYY143 pKa = 10.64FIAQSWQAIIAKK155 pKa = 10.2LPFSTLNII163 pKa = 3.97
MM1 pKa = 7.66DD2 pKa = 4.87HH3 pKa = 7.31ALTRR7 pKa = 11.84EE8 pKa = 4.04EE9 pKa = 4.51SVKK12 pKa = 10.73LMQLLKK18 pKa = 10.99LPMEE22 pKa = 4.41QYY24 pKa = 11.84GNFPLMRR31 pKa = 11.84RR32 pKa = 11.84AFLEE36 pKa = 3.98QCKK39 pKa = 10.09VLHH42 pKa = 6.76PDD44 pKa = 2.8KK45 pKa = 11.29GGNAEE50 pKa = 4.2LAKK53 pKa = 10.44EE54 pKa = 4.26LISLYY59 pKa = 10.42RR60 pKa = 11.84KK61 pKa = 10.03LEE63 pKa = 3.91AVIPPLNPNDD73 pKa = 3.78SFTTEE78 pKa = 3.77QVGVSDD84 pKa = 3.96SFVFYY89 pKa = 11.01LRR91 pKa = 11.84DD92 pKa = 3.14WSSCLKK98 pKa = 9.28GTEE101 pKa = 3.73VCICLFCLLRR111 pKa = 11.84DD112 pKa = 3.36SHH114 pKa = 6.52RR115 pKa = 11.84RR116 pKa = 11.84RR117 pKa = 11.84GSKK120 pKa = 8.99PKK122 pKa = 10.03CWGSCFCFEE131 pKa = 5.91CYY133 pKa = 9.67IAWFGLEE140 pKa = 3.75YY141 pKa = 10.01TYY143 pKa = 10.64FIAQSWQAIIAKK155 pKa = 10.2LPFSTLNII163 pKa = 3.97
Molecular weight: 18.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1497 |
163 |
654 |
374.3 |
42.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.279 ± 1.714 | 2.138 ± 0.76 |
5.411 ± 0.565 | 7.148 ± 0.477 |
5.01 ± 0.502 | 6.279 ± 0.815 |
1.937 ± 0.203 | 5.411 ± 0.185 |
5.611 ± 1.307 | 10.955 ± 0.76 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.204 ± 0.231 | 5.478 ± 0.838 |
5.678 ± 0.799 | 3.741 ± 0.287 |
4.876 ± 0.247 | 5.611 ± 0.445 |
5.478 ± 0.679 | 5.812 ± 0.625 |
1.269 ± 0.33 | 3.674 ± 0.181 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
