
Diachasma alloeum
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Ecdysozoa; Panarthropoda; Arthropoda; Mandibulata; Pancrustacea; Hexapoda; Insecta; Dicondylia; Pterygota; Neoptera; Endopterygota; Hymenoptera; Apocrita; Parasitoida; Ichneumonoidea; Braconidae; Opiinae;
Average proteome isoelectric point is 7.26
Get precalculated fractions of proteins
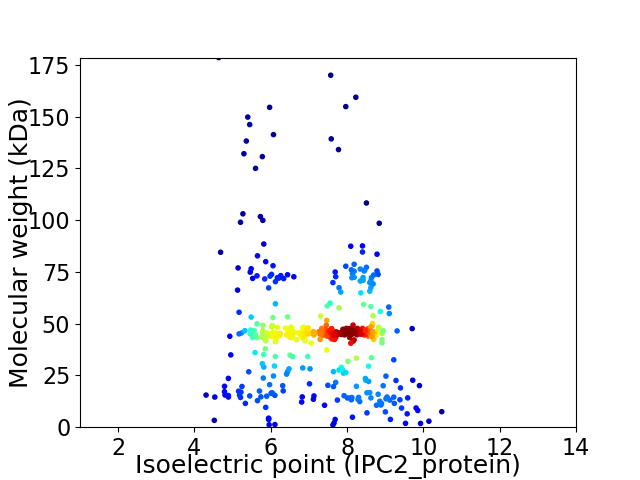
Virtual 2D-PAGE plot for 431 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
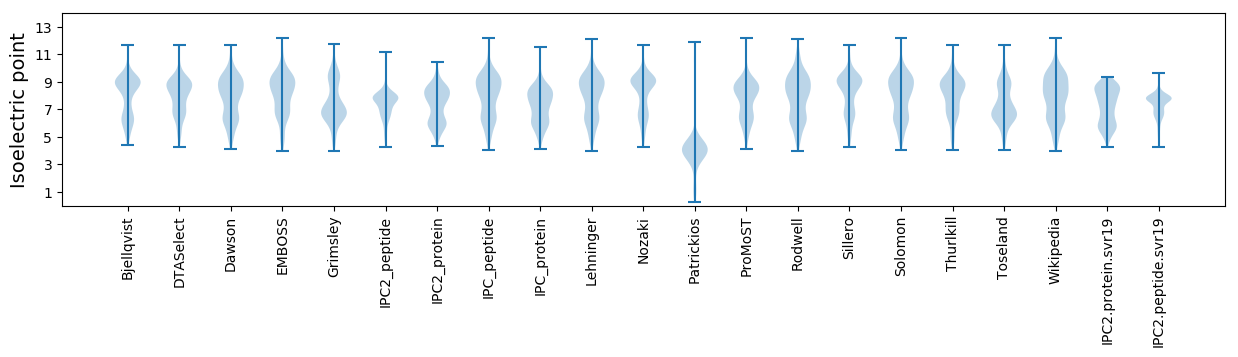
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4E0RKC0|A0A4E0RKC0_9HYME Chemosensory protein 5 OS=Diachasma alloeum OX=454923 GN=Csp5 PE=4 SV=1
MM1 pKa = 7.39ARR3 pKa = 11.84HH4 pKa = 5.5VVCCFLLGVAMQALIVSAGRR24 pKa = 11.84PDD26 pKa = 5.82FITDD30 pKa = 3.14DD31 pKa = 3.78MMAMVADD38 pKa = 4.74DD39 pKa = 4.1KK40 pKa = 11.13ARR42 pKa = 11.84CMGEE46 pKa = 3.6HH47 pKa = 6.38GTTEE51 pKa = 4.02TLIDD55 pKa = 3.9EE56 pKa = 4.89VNNGALPNDD65 pKa = 4.06RR66 pKa = 11.84ALTCYY71 pKa = 9.58MDD73 pKa = 4.11CLFAAFGVIDD83 pKa = 4.03EE84 pKa = 5.28GEE86 pKa = 4.61LEE88 pKa = 4.04VDD90 pKa = 3.51MLVGFLPDD98 pKa = 3.66HH99 pKa = 6.25MQDD102 pKa = 3.1QARR105 pKa = 11.84EE106 pKa = 3.97LLEE109 pKa = 4.75ACAKK113 pKa = 10.42QPGADD118 pKa = 3.51PCDD121 pKa = 3.44KK122 pKa = 10.61VFNIAKK128 pKa = 9.48CVQAKK133 pKa = 10.18RR134 pKa = 11.84PDD136 pKa = 3.51LWFMII141 pKa = 4.75
MM1 pKa = 7.39ARR3 pKa = 11.84HH4 pKa = 5.5VVCCFLLGVAMQALIVSAGRR24 pKa = 11.84PDD26 pKa = 5.82FITDD30 pKa = 3.14DD31 pKa = 3.78MMAMVADD38 pKa = 4.74DD39 pKa = 4.1KK40 pKa = 11.13ARR42 pKa = 11.84CMGEE46 pKa = 3.6HH47 pKa = 6.38GTTEE51 pKa = 4.02TLIDD55 pKa = 3.9EE56 pKa = 4.89VNNGALPNDD65 pKa = 4.06RR66 pKa = 11.84ALTCYY71 pKa = 9.58MDD73 pKa = 4.11CLFAAFGVIDD83 pKa = 4.03EE84 pKa = 5.28GEE86 pKa = 4.61LEE88 pKa = 4.04VDD90 pKa = 3.51MLVGFLPDD98 pKa = 3.66HH99 pKa = 6.25MQDD102 pKa = 3.1QARR105 pKa = 11.84EE106 pKa = 3.97LLEE109 pKa = 4.75ACAKK113 pKa = 10.42QPGADD118 pKa = 3.51PCDD121 pKa = 3.44KK122 pKa = 10.61VFNIAKK128 pKa = 9.48CVQAKK133 pKa = 10.18RR134 pKa = 11.84PDD136 pKa = 3.51LWFMII141 pKa = 4.75
Molecular weight: 15.52 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4E0S3T2|A0A4E0S3T2_9HYME DNA helicase OS=Diachasma alloeum OX=454923 GN=RECQ4 PE=3 SV=1
MM1 pKa = 7.56RR2 pKa = 11.84RR3 pKa = 11.84SDD5 pKa = 3.51DD6 pKa = 3.15RR7 pKa = 11.84SRR9 pKa = 11.84EE10 pKa = 3.91YY11 pKa = 10.7RR12 pKa = 11.84DD13 pKa = 3.23SRR15 pKa = 11.84DD16 pKa = 3.48YY17 pKa = 11.43LPEE20 pKa = 4.71DD21 pKa = 3.41RR22 pKa = 11.84LRR24 pKa = 11.84EE25 pKa = 4.08LEE27 pKa = 3.96RR28 pKa = 11.84KK29 pKa = 8.67RR30 pKa = 11.84RR31 pKa = 11.84EE32 pKa = 3.56WRR34 pKa = 11.84IEE36 pKa = 3.59QEE38 pKa = 3.69KK39 pKa = 10.54RR40 pKa = 11.84RR41 pKa = 11.84EE42 pKa = 3.94HH43 pKa = 6.43EE44 pKa = 3.75KK45 pKa = 10.61RR46 pKa = 11.84KK47 pKa = 10.28QKK49 pKa = 9.99MIQEE53 pKa = 4.45WEE55 pKa = 3.94AKK57 pKa = 9.37RR58 pKa = 11.84ARR60 pKa = 11.84EE61 pKa = 4.5LEE63 pKa = 4.06GKK65 pKa = 7.43NQRR68 pKa = 11.84RR69 pKa = 11.84RR70 pKa = 11.84SKK72 pKa = 10.52SRR74 pKa = 11.84SRR76 pKa = 11.84SASPGPSRR84 pKa = 11.84RR85 pKa = 11.84RR86 pKa = 11.84GRR88 pKa = 11.84SRR90 pKa = 11.84SVEE93 pKa = 4.14KK94 pKa = 10.95NSTRR98 pKa = 11.84ASTKK102 pKa = 9.46VPVMSEE108 pKa = 4.1RR109 pKa = 11.84FDD111 pKa = 3.7SASSSNTPLFKK122 pKa = 10.82GMEE125 pKa = 4.1GSKK128 pKa = 10.28ISVSEE133 pKa = 3.93LKK135 pKa = 10.38KK136 pKa = 10.48IKK138 pKa = 10.82VNIQRR143 pKa = 11.84DD144 pKa = 3.29ISKK147 pKa = 10.93AEE149 pKa = 3.95EE150 pKa = 3.93TSEE153 pKa = 4.09LLRR156 pKa = 11.84DD157 pKa = 3.31ITSPDD162 pKa = 3.47EE163 pKa = 3.94IVLKK167 pKa = 10.49RR168 pKa = 11.84RR169 pKa = 11.84EE170 pKa = 4.37GEE172 pKa = 3.8GSKK175 pKa = 10.56PIFEE179 pKa = 4.22RR180 pKa = 11.84EE181 pKa = 3.74EE182 pKa = 4.26LKK184 pKa = 10.24TKK186 pKa = 10.25EE187 pKa = 4.37SNNVDD192 pKa = 3.2VPEE195 pKa = 4.21RR196 pKa = 11.84RR197 pKa = 11.84TVVALDD203 pKa = 3.41EE204 pKa = 4.74FEE206 pKa = 4.54TNKK209 pKa = 9.8NRR211 pKa = 11.84STEE214 pKa = 3.8RR215 pKa = 11.84RR216 pKa = 11.84SRR218 pKa = 11.84SLSSGRR224 pKa = 11.84RR225 pKa = 11.84TSHH228 pKa = 5.62SSRR231 pKa = 11.84RR232 pKa = 11.84ASRR235 pKa = 11.84HH236 pKa = 5.04DD237 pKa = 3.62AASKK241 pKa = 10.67SYY243 pKa = 9.31TEE245 pKa = 3.87RR246 pKa = 11.84HH247 pKa = 6.17DD248 pKa = 4.3RR249 pKa = 11.84GHH251 pKa = 6.48SYY253 pKa = 11.46DD254 pKa = 4.42RR255 pKa = 11.84GSTTNDD261 pKa = 2.82RR262 pKa = 11.84YY263 pKa = 10.59HH264 pKa = 5.87EE265 pKa = 4.21RR266 pKa = 11.84KK267 pKa = 9.78YY268 pKa = 10.29PSSRR272 pKa = 11.84KK273 pKa = 9.36SSPEE277 pKa = 3.3RR278 pKa = 11.84VRR280 pKa = 11.84DD281 pKa = 3.53SRR283 pKa = 11.84SRR285 pKa = 11.84RR286 pKa = 11.84SQDD289 pKa = 2.57RR290 pKa = 11.84NARR293 pKa = 11.84AGSHH297 pKa = 6.62RR298 pKa = 11.84DD299 pKa = 3.38STCRR303 pKa = 11.84EE304 pKa = 3.86YY305 pKa = 11.05QDD307 pKa = 3.63YY308 pKa = 11.06DD309 pKa = 3.95SYY311 pKa = 11.32SHH313 pKa = 6.83PRR315 pKa = 11.84RR316 pKa = 11.84HH317 pKa = 5.65EE318 pKa = 3.75VDD320 pKa = 3.23RR321 pKa = 11.84RR322 pKa = 11.84PAALSYY328 pKa = 11.04AEE330 pKa = 5.41PITFPMYY337 pKa = 10.11YY338 pKa = 10.22DD339 pKa = 3.4NLVRR343 pKa = 11.84PMMMDD348 pKa = 3.7PMMMMRR354 pKa = 11.84TPMPMMRR361 pKa = 11.84GRR363 pKa = 11.84VPAMIPTFRR372 pKa = 11.84PPFRR376 pKa = 11.84PRR378 pKa = 11.84FMPSEE383 pKa = 4.24MFRR386 pKa = 11.84LNGPPTQRR394 pKa = 11.84YY395 pKa = 9.03GRR397 pKa = 11.84MFPP400 pKa = 3.75
MM1 pKa = 7.56RR2 pKa = 11.84RR3 pKa = 11.84SDD5 pKa = 3.51DD6 pKa = 3.15RR7 pKa = 11.84SRR9 pKa = 11.84EE10 pKa = 3.91YY11 pKa = 10.7RR12 pKa = 11.84DD13 pKa = 3.23SRR15 pKa = 11.84DD16 pKa = 3.48YY17 pKa = 11.43LPEE20 pKa = 4.71DD21 pKa = 3.41RR22 pKa = 11.84LRR24 pKa = 11.84EE25 pKa = 4.08LEE27 pKa = 3.96RR28 pKa = 11.84KK29 pKa = 8.67RR30 pKa = 11.84RR31 pKa = 11.84EE32 pKa = 3.56WRR34 pKa = 11.84IEE36 pKa = 3.59QEE38 pKa = 3.69KK39 pKa = 10.54RR40 pKa = 11.84RR41 pKa = 11.84EE42 pKa = 3.94HH43 pKa = 6.43EE44 pKa = 3.75KK45 pKa = 10.61RR46 pKa = 11.84KK47 pKa = 10.28QKK49 pKa = 9.99MIQEE53 pKa = 4.45WEE55 pKa = 3.94AKK57 pKa = 9.37RR58 pKa = 11.84ARR60 pKa = 11.84EE61 pKa = 4.5LEE63 pKa = 4.06GKK65 pKa = 7.43NQRR68 pKa = 11.84RR69 pKa = 11.84RR70 pKa = 11.84SKK72 pKa = 10.52SRR74 pKa = 11.84SRR76 pKa = 11.84SASPGPSRR84 pKa = 11.84RR85 pKa = 11.84RR86 pKa = 11.84GRR88 pKa = 11.84SRR90 pKa = 11.84SVEE93 pKa = 4.14KK94 pKa = 10.95NSTRR98 pKa = 11.84ASTKK102 pKa = 9.46VPVMSEE108 pKa = 4.1RR109 pKa = 11.84FDD111 pKa = 3.7SASSSNTPLFKK122 pKa = 10.82GMEE125 pKa = 4.1GSKK128 pKa = 10.28ISVSEE133 pKa = 3.93LKK135 pKa = 10.38KK136 pKa = 10.48IKK138 pKa = 10.82VNIQRR143 pKa = 11.84DD144 pKa = 3.29ISKK147 pKa = 10.93AEE149 pKa = 3.95EE150 pKa = 3.93TSEE153 pKa = 4.09LLRR156 pKa = 11.84DD157 pKa = 3.31ITSPDD162 pKa = 3.47EE163 pKa = 3.94IVLKK167 pKa = 10.49RR168 pKa = 11.84RR169 pKa = 11.84EE170 pKa = 4.37GEE172 pKa = 3.8GSKK175 pKa = 10.56PIFEE179 pKa = 4.22RR180 pKa = 11.84EE181 pKa = 3.74EE182 pKa = 4.26LKK184 pKa = 10.24TKK186 pKa = 10.25EE187 pKa = 4.37SNNVDD192 pKa = 3.2VPEE195 pKa = 4.21RR196 pKa = 11.84RR197 pKa = 11.84TVVALDD203 pKa = 3.41EE204 pKa = 4.74FEE206 pKa = 4.54TNKK209 pKa = 9.8NRR211 pKa = 11.84STEE214 pKa = 3.8RR215 pKa = 11.84RR216 pKa = 11.84SRR218 pKa = 11.84SLSSGRR224 pKa = 11.84RR225 pKa = 11.84TSHH228 pKa = 5.62SSRR231 pKa = 11.84RR232 pKa = 11.84ASRR235 pKa = 11.84HH236 pKa = 5.04DD237 pKa = 3.62AASKK241 pKa = 10.67SYY243 pKa = 9.31TEE245 pKa = 3.87RR246 pKa = 11.84HH247 pKa = 6.17DD248 pKa = 4.3RR249 pKa = 11.84GHH251 pKa = 6.48SYY253 pKa = 11.46DD254 pKa = 4.42RR255 pKa = 11.84GSTTNDD261 pKa = 2.82RR262 pKa = 11.84YY263 pKa = 10.59HH264 pKa = 5.87EE265 pKa = 4.21RR266 pKa = 11.84KK267 pKa = 9.78YY268 pKa = 10.29PSSRR272 pKa = 11.84KK273 pKa = 9.36SSPEE277 pKa = 3.3RR278 pKa = 11.84VRR280 pKa = 11.84DD281 pKa = 3.53SRR283 pKa = 11.84SRR285 pKa = 11.84RR286 pKa = 11.84SQDD289 pKa = 2.57RR290 pKa = 11.84NARR293 pKa = 11.84AGSHH297 pKa = 6.62RR298 pKa = 11.84DD299 pKa = 3.38STCRR303 pKa = 11.84EE304 pKa = 3.86YY305 pKa = 11.05QDD307 pKa = 3.63YY308 pKa = 11.06DD309 pKa = 3.95SYY311 pKa = 11.32SHH313 pKa = 6.83PRR315 pKa = 11.84RR316 pKa = 11.84HH317 pKa = 5.65EE318 pKa = 3.75VDD320 pKa = 3.23RR321 pKa = 11.84RR322 pKa = 11.84PAALSYY328 pKa = 11.04AEE330 pKa = 5.41PITFPMYY337 pKa = 10.11YY338 pKa = 10.22DD339 pKa = 3.4NLVRR343 pKa = 11.84PMMMDD348 pKa = 3.7PMMMMRR354 pKa = 11.84TPMPMMRR361 pKa = 11.84GRR363 pKa = 11.84VPAMIPTFRR372 pKa = 11.84PPFRR376 pKa = 11.84PRR378 pKa = 11.84FMPSEE383 pKa = 4.24MFRR386 pKa = 11.84LNGPPTQRR394 pKa = 11.84YY395 pKa = 9.03GRR397 pKa = 11.84MFPP400 pKa = 3.75
Molecular weight: 47.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
172772 |
9 |
1591 |
400.9 |
45.81 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.076 ± 0.085 | 2.084 ± 0.054 |
4.303 ± 0.084 | 5.659 ± 0.155 |
5.563 ± 0.111 | 4.81 ± 0.066 |
2.231 ± 0.038 | 8.086 ± 0.118 |
5.564 ± 0.118 | 10.539 ± 0.111 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.248 ± 0.05 | 4.177 ± 0.068 |
3.642 ± 0.086 | 3.459 ± 0.062 |
5.172 ± 0.093 | 7.291 ± 0.083 |
5.952 ± 0.058 | 6.699 ± 0.077 |
1.466 ± 0.039 | 3.979 ± 0.067 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
