
Geodermatophilus sp. TF02-6
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Geodermatophilales; Geodermatophilaceae; Geodermatophilus; unclassified Geodermatophilus
Average proteome isoelectric point is 6.28
Get precalculated fractions of proteins
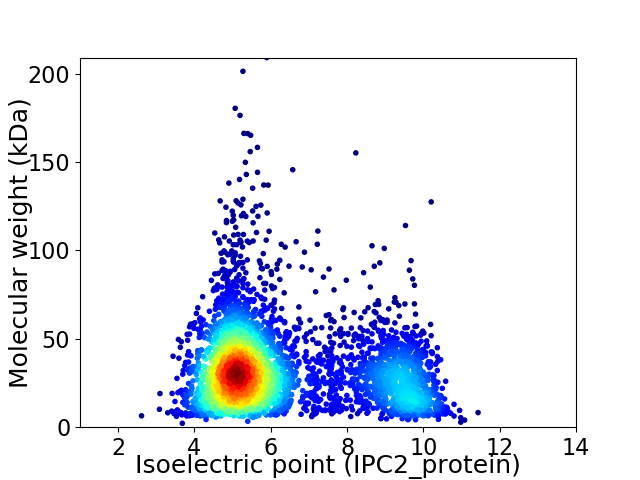
Virtual 2D-PAGE plot for 4244 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A367A3L8|A0A367A3L8_9ACTN TerC family protein OS=Geodermatophilus sp. TF02-6 OX=2250575 GN=DQ238_03350 PE=3 SV=1
MM1 pKa = 7.6AATPPRR7 pKa = 11.84RR8 pKa = 11.84TLAALLAAVALLLVSGCSIVVPGRR32 pKa = 11.84PSAAQPPTDD41 pKa = 3.73DD42 pKa = 3.59VAPGEE47 pKa = 4.14LAVAGATGGPIDD59 pKa = 3.82TLARR63 pKa = 11.84NALTDD68 pKa = 4.66LEE70 pKa = 4.96TYY72 pKa = 8.72WAGQFPDD79 pKa = 4.15VFGQPFQPLQGGYY92 pKa = 10.53FSVDD96 pKa = 3.44PGNVAPGEE104 pKa = 4.13FPQGIGCGADD114 pKa = 3.38PLDD117 pKa = 3.7VEE119 pKa = 5.82GNAFYY124 pKa = 10.7CQASDD129 pKa = 4.2APNSDD134 pKa = 3.73SISYY138 pKa = 10.51DD139 pKa = 3.0RR140 pKa = 11.84AFLQEE145 pKa = 4.66LADD148 pKa = 3.44QYY150 pKa = 11.9GRR152 pKa = 11.84FIPALVMAHH161 pKa = 6.2EE162 pKa = 5.31FGHH165 pKa = 6.69AVQARR170 pKa = 11.84VGYY173 pKa = 8.38PDD175 pKa = 3.67YY176 pKa = 10.82SISVEE181 pKa = 4.12TQADD185 pKa = 4.0CFAGAWTAWVADD197 pKa = 3.98GQAEE201 pKa = 4.23HH202 pKa = 6.15SQIRR206 pKa = 11.84APEE209 pKa = 4.1LDD211 pKa = 3.48EE212 pKa = 4.08VLRR215 pKa = 11.84GYY217 pKa = 11.13LLLRR221 pKa = 11.84DD222 pKa = 3.92PVGTSINTEE231 pKa = 4.01AAHH234 pKa = 6.51GSYY237 pKa = 10.46FDD239 pKa = 3.78RR240 pKa = 11.84VSAFQEE246 pKa = 4.33GFEE249 pKa = 4.8AGPTACRR256 pKa = 11.84DD257 pKa = 3.43DD258 pKa = 4.05FTAQRR263 pKa = 11.84PYY265 pKa = 10.15TQGAFQDD272 pKa = 3.78PSEE275 pKa = 4.33ARR277 pKa = 11.84TGGNSSFRR285 pKa = 11.84EE286 pKa = 4.18AQDD289 pKa = 2.64IAATVLPEE297 pKa = 3.83FWNRR301 pKa = 11.84AFTEE305 pKa = 4.2VFDD308 pKa = 4.31ATFSPPTLEE317 pKa = 4.72PFTGTAPSCAPDD329 pKa = 4.37DD330 pKa = 4.52LDD332 pKa = 5.81LVFCADD338 pKa = 4.01EE339 pKa = 4.23NLVGYY344 pKa = 9.92DD345 pKa = 3.99EE346 pKa = 4.83QDD348 pKa = 3.34LAAPAYY354 pKa = 9.13QQIGDD359 pKa = 4.07YY360 pKa = 10.96AVVTAASLPYY370 pKa = 9.28AQSGRR375 pKa = 11.84EE376 pKa = 3.75QLGRR380 pKa = 11.84STDD383 pKa = 3.42DD384 pKa = 3.02EE385 pKa = 4.23AAIRR389 pKa = 11.84SAVCLTGWFSAAFFDD404 pKa = 5.63GALQSARR411 pKa = 11.84ISPGDD416 pKa = 3.3IDD418 pKa = 4.53EE419 pKa = 4.65SVQFLLEE426 pKa = 4.13YY427 pKa = 9.56GTDD430 pKa = 3.41PSVFPDD436 pKa = 2.99VDD438 pKa = 3.65LTGFQLVDD446 pKa = 3.55LFRR449 pKa = 11.84NGFFDD454 pKa = 4.68GAAACDD460 pKa = 3.64VGVV463 pKa = 3.59
MM1 pKa = 7.6AATPPRR7 pKa = 11.84RR8 pKa = 11.84TLAALLAAVALLLVSGCSIVVPGRR32 pKa = 11.84PSAAQPPTDD41 pKa = 3.73DD42 pKa = 3.59VAPGEE47 pKa = 4.14LAVAGATGGPIDD59 pKa = 3.82TLARR63 pKa = 11.84NALTDD68 pKa = 4.66LEE70 pKa = 4.96TYY72 pKa = 8.72WAGQFPDD79 pKa = 4.15VFGQPFQPLQGGYY92 pKa = 10.53FSVDD96 pKa = 3.44PGNVAPGEE104 pKa = 4.13FPQGIGCGADD114 pKa = 3.38PLDD117 pKa = 3.7VEE119 pKa = 5.82GNAFYY124 pKa = 10.7CQASDD129 pKa = 4.2APNSDD134 pKa = 3.73SISYY138 pKa = 10.51DD139 pKa = 3.0RR140 pKa = 11.84AFLQEE145 pKa = 4.66LADD148 pKa = 3.44QYY150 pKa = 11.9GRR152 pKa = 11.84FIPALVMAHH161 pKa = 6.2EE162 pKa = 5.31FGHH165 pKa = 6.69AVQARR170 pKa = 11.84VGYY173 pKa = 8.38PDD175 pKa = 3.67YY176 pKa = 10.82SISVEE181 pKa = 4.12TQADD185 pKa = 4.0CFAGAWTAWVADD197 pKa = 3.98GQAEE201 pKa = 4.23HH202 pKa = 6.15SQIRR206 pKa = 11.84APEE209 pKa = 4.1LDD211 pKa = 3.48EE212 pKa = 4.08VLRR215 pKa = 11.84GYY217 pKa = 11.13LLLRR221 pKa = 11.84DD222 pKa = 3.92PVGTSINTEE231 pKa = 4.01AAHH234 pKa = 6.51GSYY237 pKa = 10.46FDD239 pKa = 3.78RR240 pKa = 11.84VSAFQEE246 pKa = 4.33GFEE249 pKa = 4.8AGPTACRR256 pKa = 11.84DD257 pKa = 3.43DD258 pKa = 4.05FTAQRR263 pKa = 11.84PYY265 pKa = 10.15TQGAFQDD272 pKa = 3.78PSEE275 pKa = 4.33ARR277 pKa = 11.84TGGNSSFRR285 pKa = 11.84EE286 pKa = 4.18AQDD289 pKa = 2.64IAATVLPEE297 pKa = 3.83FWNRR301 pKa = 11.84AFTEE305 pKa = 4.2VFDD308 pKa = 4.31ATFSPPTLEE317 pKa = 4.72PFTGTAPSCAPDD329 pKa = 4.37DD330 pKa = 4.52LDD332 pKa = 5.81LVFCADD338 pKa = 4.01EE339 pKa = 4.23NLVGYY344 pKa = 9.92DD345 pKa = 3.99EE346 pKa = 4.83QDD348 pKa = 3.34LAAPAYY354 pKa = 9.13QQIGDD359 pKa = 4.07YY360 pKa = 10.96AVVTAASLPYY370 pKa = 9.28AQSGRR375 pKa = 11.84EE376 pKa = 3.75QLGRR380 pKa = 11.84STDD383 pKa = 3.42DD384 pKa = 3.02EE385 pKa = 4.23AAIRR389 pKa = 11.84SAVCLTGWFSAAFFDD404 pKa = 5.63GALQSARR411 pKa = 11.84ISPGDD416 pKa = 3.3IDD418 pKa = 4.53EE419 pKa = 4.65SVQFLLEE426 pKa = 4.13YY427 pKa = 9.56GTDD430 pKa = 3.41PSVFPDD436 pKa = 2.99VDD438 pKa = 3.65LTGFQLVDD446 pKa = 3.55LFRR449 pKa = 11.84NGFFDD454 pKa = 4.68GAAACDD460 pKa = 3.64VGVV463 pKa = 3.59
Molecular weight: 49.19 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A366ZDI2|A0A366ZDI2_9ACTN VWFA domain-containing protein OS=Geodermatophilus sp. TF02-6 OX=2250575 GN=DQ238_20375 PE=4 SV=1
MM1 pKa = 7.3SGRR4 pKa = 11.84RR5 pKa = 11.84GSGRR9 pKa = 11.84GMAGWAMSGRR19 pKa = 11.84GLSGRR24 pKa = 11.84GGSGRR29 pKa = 11.84GMAGRR34 pKa = 11.84GMAGWAMSGRR44 pKa = 11.84GASAGRR50 pKa = 11.84GHH52 pKa = 6.91GRR54 pKa = 11.84PTSRR58 pKa = 11.84GMRR61 pKa = 11.84RR62 pKa = 11.84GRR64 pKa = 11.84PTTAASGLGGVLSGLLRR81 pKa = 11.84RR82 pKa = 11.84RR83 pKa = 3.65
MM1 pKa = 7.3SGRR4 pKa = 11.84RR5 pKa = 11.84GSGRR9 pKa = 11.84GMAGWAMSGRR19 pKa = 11.84GLSGRR24 pKa = 11.84GGSGRR29 pKa = 11.84GMAGRR34 pKa = 11.84GMAGWAMSGRR44 pKa = 11.84GASAGRR50 pKa = 11.84GHH52 pKa = 6.91GRR54 pKa = 11.84PTSRR58 pKa = 11.84GMRR61 pKa = 11.84RR62 pKa = 11.84GRR64 pKa = 11.84PTTAASGLGGVLSGLLRR81 pKa = 11.84RR82 pKa = 11.84RR83 pKa = 3.65
Molecular weight: 8.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1348583 |
20 |
1980 |
317.8 |
33.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.467 ± 0.067 | 0.736 ± 0.01 |
6.251 ± 0.027 | 5.48 ± 0.039 |
2.5 ± 0.021 | 9.663 ± 0.036 |
2.081 ± 0.018 | 2.501 ± 0.025 |
1.197 ± 0.018 | 10.611 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.499 ± 0.015 | 1.334 ± 0.015 |
6.439 ± 0.033 | 2.677 ± 0.021 |
8.682 ± 0.038 | 4.608 ± 0.025 |
5.925 ± 0.029 | 10.098 ± 0.038 |
1.48 ± 0.017 | 1.769 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
