
Sphingosinicella microcystinivorans
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingosinicellaceae; Sphingosinicella
Average proteome isoelectric point is 6.44
Get precalculated fractions of proteins
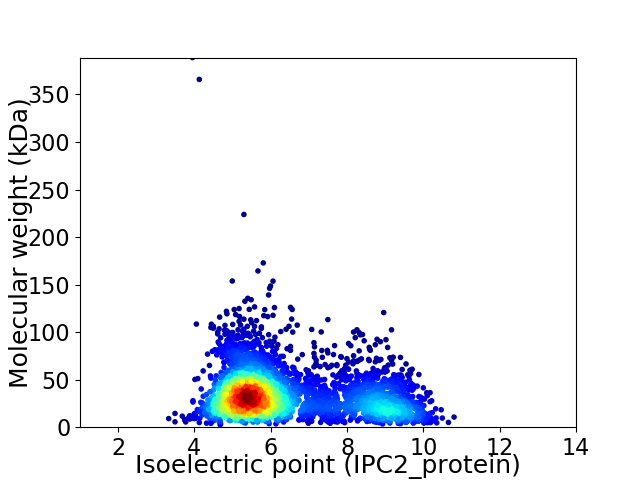
Virtual 2D-PAGE plot for 3645 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
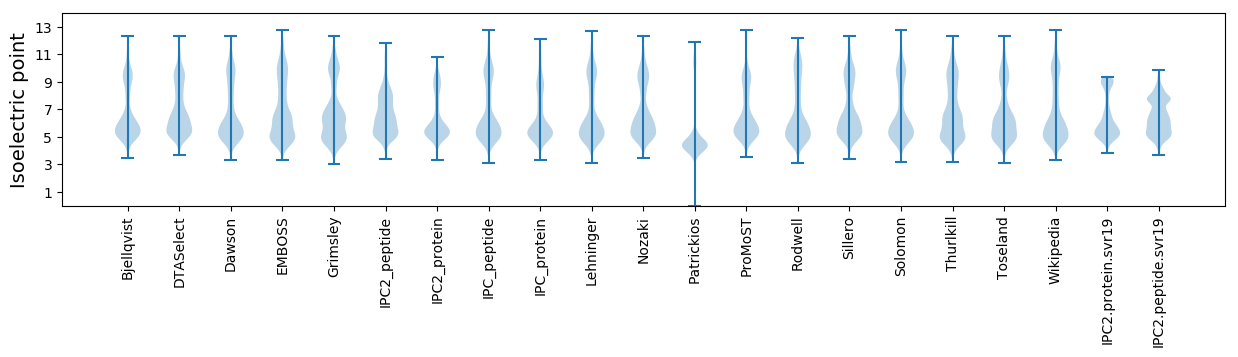
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A495RWP5|A0A495RWP5_SPHMI Dihydrolipoyl dehydrogenase OS=Sphingosinicella microcystinivorans OX=335406 GN=DFR51_1562 PE=3 SV=1
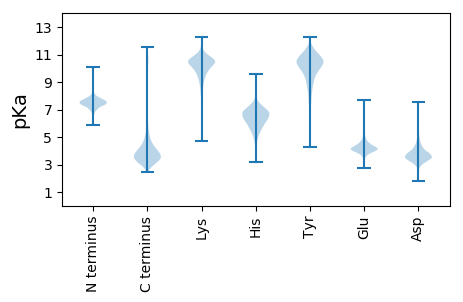
MM1 pKa = 7.54KK2 pKa = 10.39KK3 pKa = 10.3GIRR6 pKa = 11.84SRR8 pKa = 11.84AISTTVLAAATLVGSLAVAAPAFAQDD34 pKa = 4.73AEE36 pKa = 4.44ATSDD40 pKa = 3.38WEE42 pKa = 3.9ISGNAGIFSQYY53 pKa = 7.31RR54 pKa = 11.84WRR56 pKa = 11.84GISLSDD62 pKa = 3.63EE63 pKa = 3.98DD64 pKa = 4.16VAFQGGIDD72 pKa = 3.76AAHH75 pKa = 6.47SSGFYY80 pKa = 9.94VGTWGSSLAGYY91 pKa = 10.1GSYY94 pKa = 10.66GGSNTEE100 pKa = 3.29IDD102 pKa = 4.04VYY104 pKa = 11.18AGYY107 pKa = 10.61AGEE110 pKa = 4.68AGGFGYY116 pKa = 10.44DD117 pKa = 5.08IGAIWYY123 pKa = 7.2LYY125 pKa = 9.63PGTSGTDD132 pKa = 3.26VVEE135 pKa = 4.5ITASLSKK142 pKa = 10.61EE143 pKa = 4.04LGPVGASVGVAYY155 pKa = 10.16APKK158 pKa = 9.65QDD160 pKa = 4.27SLSSDD165 pKa = 3.3DD166 pKa = 4.88SFYY169 pKa = 11.24LYY171 pKa = 10.19TDD173 pKa = 3.63WSAAIPNTPVSLNAHH188 pKa = 6.94LGYY191 pKa = 9.41TDD193 pKa = 3.95GSLSIDD199 pKa = 3.3RR200 pKa = 11.84GFSTDD205 pKa = 3.51GDD207 pKa = 4.04YY208 pKa = 11.55LDD210 pKa = 3.98WSLGASVSYY219 pKa = 11.08DD220 pKa = 3.13KK221 pKa = 10.88LTLGVSYY228 pKa = 11.3VDD230 pKa = 3.53TDD232 pKa = 3.79VKK234 pKa = 11.25GGDD237 pKa = 3.43PSINKK242 pKa = 8.26ITDD245 pKa = 3.13AAIIVSLTAAFF256 pKa = 3.92
MM1 pKa = 7.54KK2 pKa = 10.39KK3 pKa = 10.3GIRR6 pKa = 11.84SRR8 pKa = 11.84AISTTVLAAATLVGSLAVAAPAFAQDD34 pKa = 4.73AEE36 pKa = 4.44ATSDD40 pKa = 3.38WEE42 pKa = 3.9ISGNAGIFSQYY53 pKa = 7.31RR54 pKa = 11.84WRR56 pKa = 11.84GISLSDD62 pKa = 3.63EE63 pKa = 3.98DD64 pKa = 4.16VAFQGGIDD72 pKa = 3.76AAHH75 pKa = 6.47SSGFYY80 pKa = 9.94VGTWGSSLAGYY91 pKa = 10.1GSYY94 pKa = 10.66GGSNTEE100 pKa = 3.29IDD102 pKa = 4.04VYY104 pKa = 11.18AGYY107 pKa = 10.61AGEE110 pKa = 4.68AGGFGYY116 pKa = 10.44DD117 pKa = 5.08IGAIWYY123 pKa = 7.2LYY125 pKa = 9.63PGTSGTDD132 pKa = 3.26VVEE135 pKa = 4.5ITASLSKK142 pKa = 10.61EE143 pKa = 4.04LGPVGASVGVAYY155 pKa = 10.16APKK158 pKa = 9.65QDD160 pKa = 4.27SLSSDD165 pKa = 3.3DD166 pKa = 4.88SFYY169 pKa = 11.24LYY171 pKa = 10.19TDD173 pKa = 3.63WSAAIPNTPVSLNAHH188 pKa = 6.94LGYY191 pKa = 9.41TDD193 pKa = 3.95GSLSIDD199 pKa = 3.3RR200 pKa = 11.84GFSTDD205 pKa = 3.51GDD207 pKa = 4.04YY208 pKa = 11.55LDD210 pKa = 3.98WSLGASVSYY219 pKa = 11.08DD220 pKa = 3.13KK221 pKa = 10.88LTLGVSYY228 pKa = 11.3VDD230 pKa = 3.53TDD232 pKa = 3.79VKK234 pKa = 11.25GGDD237 pKa = 3.43PSINKK242 pKa = 8.26ITDD245 pKa = 3.13AAIIVSLTAAFF256 pKa = 3.92
Molecular weight: 26.49 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A495RDJ9|A0A495RDJ9_SPHMI tRNA uridine 5-carboxymethylaminomethyl modification enzyme MnmG OS=Sphingosinicella microcystinivorans OX=335406 GN=mnmG PE=3 SV=1
MM1 pKa = 7.63KK2 pKa = 9.74PPPHH6 pKa = 6.84RR7 pKa = 11.84ASDD10 pKa = 3.47PGLRR14 pKa = 11.84RR15 pKa = 11.84QQFGHH20 pKa = 5.7VVWVHH25 pKa = 6.47CPQCDD30 pKa = 3.76GPAKK34 pKa = 10.5HH35 pKa = 6.27NALGVKK41 pKa = 9.81CIRR44 pKa = 11.84CGYY47 pKa = 6.57MTIPVIKK54 pKa = 9.49PAAVQWARR62 pKa = 11.84ITISDD67 pKa = 4.5PRR69 pKa = 11.84CSHH72 pKa = 6.45CRR74 pKa = 11.84NPLPDD79 pKa = 3.3TSRR82 pKa = 11.84PLARR86 pKa = 11.84GGDD89 pKa = 3.7EE90 pKa = 3.77NRR92 pKa = 11.84PVVRR96 pKa = 11.84VRR98 pKa = 11.84CPHH101 pKa = 6.24CAKK104 pKa = 10.15TMAYY108 pKa = 8.24PARR111 pKa = 11.84AWSAPSRR118 pKa = 11.84ASQQQPRR125 pKa = 11.84RR126 pKa = 11.84LKK128 pKa = 10.34PYY130 pKa = 9.07LTAQVAGNLLMVDD143 pKa = 3.97NLAHH147 pKa = 6.94LSALEE152 pKa = 4.34TYY154 pKa = 10.53LGAALRR160 pKa = 11.84EE161 pKa = 4.07RR162 pKa = 11.84GPVRR166 pKa = 11.84GLTMMARR173 pKa = 11.84LPAWMKK179 pKa = 10.74SSTNRR184 pKa = 11.84AKK186 pKa = 10.47ILRR189 pKa = 11.84SLRR192 pKa = 11.84QLADD196 pKa = 3.55RR197 pKa = 11.84ASKK200 pKa = 10.87DD201 pKa = 3.8GIDD204 pKa = 3.58EE205 pKa = 4.14
MM1 pKa = 7.63KK2 pKa = 9.74PPPHH6 pKa = 6.84RR7 pKa = 11.84ASDD10 pKa = 3.47PGLRR14 pKa = 11.84RR15 pKa = 11.84QQFGHH20 pKa = 5.7VVWVHH25 pKa = 6.47CPQCDD30 pKa = 3.76GPAKK34 pKa = 10.5HH35 pKa = 6.27NALGVKK41 pKa = 9.81CIRR44 pKa = 11.84CGYY47 pKa = 6.57MTIPVIKK54 pKa = 9.49PAAVQWARR62 pKa = 11.84ITISDD67 pKa = 4.5PRR69 pKa = 11.84CSHH72 pKa = 6.45CRR74 pKa = 11.84NPLPDD79 pKa = 3.3TSRR82 pKa = 11.84PLARR86 pKa = 11.84GGDD89 pKa = 3.7EE90 pKa = 3.77NRR92 pKa = 11.84PVVRR96 pKa = 11.84VRR98 pKa = 11.84CPHH101 pKa = 6.24CAKK104 pKa = 10.15TMAYY108 pKa = 8.24PARR111 pKa = 11.84AWSAPSRR118 pKa = 11.84ASQQQPRR125 pKa = 11.84RR126 pKa = 11.84LKK128 pKa = 10.34PYY130 pKa = 9.07LTAQVAGNLLMVDD143 pKa = 3.97NLAHH147 pKa = 6.94LSALEE152 pKa = 4.34TYY154 pKa = 10.53LGAALRR160 pKa = 11.84EE161 pKa = 4.07RR162 pKa = 11.84GPVRR166 pKa = 11.84GLTMMARR173 pKa = 11.84LPAWMKK179 pKa = 10.74SSTNRR184 pKa = 11.84AKK186 pKa = 10.47ILRR189 pKa = 11.84SLRR192 pKa = 11.84QLADD196 pKa = 3.55RR197 pKa = 11.84ASKK200 pKa = 10.87DD201 pKa = 3.8GIDD204 pKa = 3.58EE205 pKa = 4.14
Molecular weight: 22.76 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1206989 |
25 |
3982 |
331.1 |
35.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.27 ± 0.061 | 0.785 ± 0.013 |
5.866 ± 0.029 | 5.582 ± 0.036 |
3.69 ± 0.025 | 8.964 ± 0.052 |
1.962 ± 0.02 | 5.044 ± 0.026 |
3.124 ± 0.033 | 9.757 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.405 ± 0.022 | 2.575 ± 0.031 |
5.178 ± 0.033 | 2.759 ± 0.022 |
7.237 ± 0.045 | 5.297 ± 0.032 |
5.424 ± 0.037 | 7.355 ± 0.028 |
1.387 ± 0.017 | 2.34 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
