
Novosphingobium pentaromativorans US6-1
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Novosphingobium; Novosphingobium pentaromativorans
Average proteome isoelectric point is 6.38
Get precalculated fractions of proteins
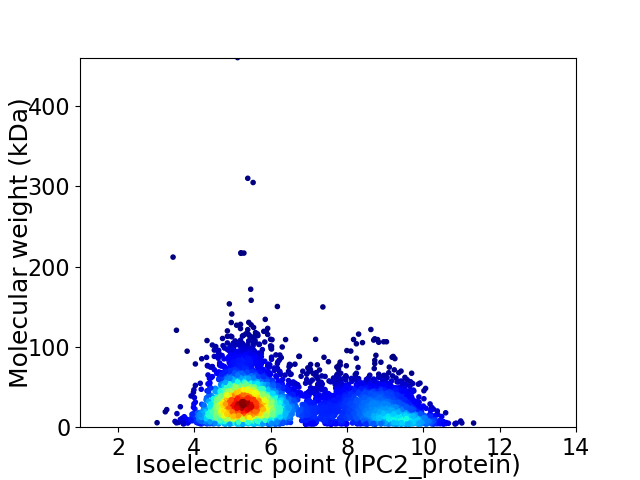
Virtual 2D-PAGE plot for 5229 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G6EJ98|G6EJ98_9SPHN Uncharacterized protein OS=Novosphingobium pentaromativorans US6-1 OX=1088721 GN=NSU_4419 PE=4 SV=1
MM1 pKa = 7.59PPAGATDD8 pKa = 3.51SFVYY12 pKa = 10.13TITDD16 pKa = 3.32GDD18 pKa = 4.4GDD20 pKa = 4.04TSTTTLTISLTDD32 pKa = 3.29SGLAASNEE40 pKa = 4.06DD41 pKa = 3.37ASVDD45 pKa = 3.65EE46 pKa = 4.54AALGIGSNPSSTAEE60 pKa = 4.18TVSGSLTDD68 pKa = 3.71NASGGAGGYY77 pKa = 8.45TYY79 pKa = 11.42ALVGSATGSHH89 pKa = 5.41GTITIDD95 pKa = 4.22PSGSWSYY102 pKa = 10.1TLTSPVDD109 pKa = 3.46GATLDD114 pKa = 3.67NGVTTEE120 pKa = 4.07NNLEE124 pKa = 4.11SFTYY128 pKa = 10.19QVTDD132 pKa = 3.57ANGNTTTSTITIDD145 pKa = 4.24VIDD148 pKa = 4.92DD149 pKa = 3.77VPTARR154 pKa = 11.84ADD156 pKa = 3.44TDD158 pKa = 4.12SVVEE162 pKa = 4.23GASTDD167 pKa = 3.66GNVLSGTGTTSGLVDD182 pKa = 3.27VLGADD187 pKa = 4.26GAAPGGAVTGVATGTDD203 pKa = 3.06TSAPVTGNLGGAGIAGTYY221 pKa = 7.35GTLILNADD229 pKa = 3.47GSYY232 pKa = 10.12TYY234 pKa = 11.23NSTANSISGNATDD247 pKa = 3.85SFVYY251 pKa = 10.1TITDD255 pKa = 3.32GDD257 pKa = 4.4GDD259 pKa = 4.04TSTTTLTISLTDD271 pKa = 3.38VSLVGDD277 pKa = 4.18DD278 pKa = 4.11EE279 pKa = 5.01SVTVYY284 pKa = 10.67EE285 pKa = 4.63KK286 pKa = 11.15ALDD289 pKa = 3.92TAQTGADD296 pKa = 3.48VAAGSITGSMPSDD309 pKa = 3.77PGEE312 pKa = 4.14TASGAVSVTGATGYY326 pKa = 10.47SIAGGSVSGNEE337 pKa = 3.95TTVVGTYY344 pKa = 7.73GTLVLNNSTGAFTYY358 pKa = 10.21TLTSPVTTTPSANDD372 pKa = 3.73GVTTALGAEE381 pKa = 4.4SFTYY385 pKa = 9.15TASDD389 pKa = 3.46ANGNTTTGTISIDD402 pKa = 3.74VVDD405 pKa = 4.38DD406 pKa = 3.45TPYY409 pKa = 11.28AVVPEE414 pKa = 5.18AIQVTNGAAPVSAPADD430 pKa = 3.73LDD432 pKa = 3.82ADD434 pKa = 3.57MSVIDD439 pKa = 4.68NYY441 pKa = 11.23GADD444 pKa = 3.41GAGTVRR450 pKa = 11.84FAPSLDD456 pKa = 3.71GADD459 pKa = 3.99SGLTHH464 pKa = 6.73NFVSITYY471 pKa = 8.28TLVSDD476 pKa = 4.52TVLEE480 pKa = 4.7AYY482 pKa = 10.22AGADD486 pKa = 3.34KK487 pKa = 10.89IFTVTLDD494 pKa = 3.71PTTATYY500 pKa = 9.29TVDD503 pKa = 3.04MDD505 pKa = 3.79GTIDD509 pKa = 4.17SIQTIDD515 pKa = 4.01FNAGGYY521 pKa = 10.3NFVGGNNSWSGFIPVGEE538 pKa = 4.46TVGSPIDD545 pKa = 4.07NNSQDD550 pKa = 4.57LLLTPSINNADD561 pKa = 4.18DD562 pKa = 3.65GTINSTANTGGIGGGASVGSNEE584 pKa = 3.97TFRR587 pKa = 11.84VDD589 pKa = 4.03FVTDD593 pKa = 3.55LRR595 pKa = 11.84GDD597 pKa = 3.87PKK599 pKa = 10.96DD600 pKa = 3.49GAGDD604 pKa = 3.9YY605 pKa = 9.02DD606 pKa = 3.92TDD608 pKa = 3.76TNRR611 pKa = 11.84DD612 pKa = 3.16HH613 pKa = 6.91MFDD616 pKa = 2.56GHH618 pKa = 5.09YY619 pKa = 9.32TVNGAVALFKK629 pKa = 10.17STNGSTVNIAAFDD642 pKa = 4.31DD643 pKa = 4.45PDD645 pKa = 4.72GNTAVGDD652 pKa = 3.84GDD654 pKa = 3.87KK655 pKa = 11.31DD656 pKa = 4.32SITGISIVWRR666 pKa = 11.84GTQYY670 pKa = 11.24VDD672 pKa = 3.7GSGDD676 pKa = 3.93PIIIPTITATNYY688 pKa = 7.78TVNGHH693 pKa = 5.33VFTVTLLGDD702 pKa = 3.77GSVNVAGVEE711 pKa = 3.98GDD713 pKa = 3.65PGSSQVGTQIAVFTGDD729 pKa = 3.17GFNSVEE735 pKa = 4.12YY736 pKa = 8.82TWAGGDD742 pKa = 3.44TFQIGDD748 pKa = 4.39FGASTLTNDD757 pKa = 4.1PVNFTIPVEE766 pKa = 4.28VVDD769 pKa = 4.37GDD771 pKa = 4.23GDD773 pKa = 3.72VSAQSNLSITAVTTTPPVALDD794 pKa = 3.64LDD796 pKa = 4.22GDD798 pKa = 4.09GVEE801 pKa = 5.61FVGLDD806 pKa = 3.01AGVTHH811 pKa = 7.96DD812 pKa = 4.2YY813 pKa = 11.45GSGLVQTAWLSADD826 pKa = 4.26DD827 pKa = 4.93GLLAHH832 pKa = 6.82DD833 pKa = 4.71TGHH836 pKa = 6.61GLDD839 pKa = 4.6IVFTDD844 pKa = 4.2DD845 pKa = 3.49AAGAEE850 pKa = 4.18TDD852 pKa = 4.38LEE854 pKa = 4.31GLRR857 pKa = 11.84LAYY860 pKa = 10.2DD861 pKa = 3.57SNGDD865 pKa = 3.67GKK867 pKa = 9.48LTAADD872 pKa = 3.59EE873 pKa = 4.72SYY875 pKa = 11.34SEE877 pKa = 4.12FGVWQDD883 pKa = 3.25ANSNGVVDD891 pKa = 4.29AGEE894 pKa = 4.36FKK896 pKa = 10.43TLAQMGITSIEE907 pKa = 4.06LTAEE911 pKa = 4.64GPGSSQADD919 pKa = 3.1GDD921 pKa = 4.83AIVHH925 pKa = 5.46ATGEE929 pKa = 4.28YY930 pKa = 8.18TMKK933 pKa = 10.67GATYY937 pKa = 10.98ALADD941 pKa = 3.59VSFATASVEE950 pKa = 4.22SQTAARR956 pKa = 11.84TAEE959 pKa = 4.02MAAITAAAAGFMMSGAAHH977 pKa = 7.38AMPLSPIAIEE987 pKa = 4.77AIAAAMQTFEE997 pKa = 4.05THH999 pKa = 5.79LPEE1002 pKa = 4.01VTVAVPEE1009 pKa = 4.32HH1010 pKa = 6.75APATPPVTFLQDD1022 pKa = 3.15GQQAHH1027 pKa = 5.91QDD1029 pKa = 3.98SEE1031 pKa = 4.78PEE1033 pKa = 4.08SHH1035 pKa = 7.36PSQHH1039 pKa = 6.62AAEE1042 pKa = 4.28QGAGHH1047 pKa = 6.82GLGDD1051 pKa = 3.82VVQHH1055 pKa = 6.05QDD1057 pKa = 3.33VSVQPLGSGEE1067 pKa = 4.13EE1068 pKa = 4.22GHH1070 pKa = 6.28VQASAAPALFGGADD1084 pKa = 3.55SGLMQALLLAAQGEE1098 pKa = 4.52QAGGSKK1104 pKa = 10.58DD1105 pKa = 3.38QGNGTGAGQGSGHH1118 pKa = 6.27GGEE1121 pKa = 4.72HH1122 pKa = 7.14GSGNLPAVQEE1132 pKa = 4.86AIAEE1136 pKa = 4.6SAGHH1140 pKa = 6.27HH1141 pKa = 6.13AVDD1144 pKa = 4.37SLIDD1148 pKa = 3.59HH1149 pKa = 7.02FAGHH1153 pKa = 5.99QGAHH1157 pKa = 6.43GAPAIAEE1164 pKa = 4.22TMGADD1169 pKa = 3.85GLDD1172 pKa = 3.55HH1173 pKa = 6.9VLGAMLAGGGLDD1185 pKa = 3.65GLHH1188 pKa = 6.73AAGLAGGFEE1197 pKa = 4.44MAAMAAAMEE1206 pKa = 4.24AHH1208 pKa = 6.04STTVAA1213 pKa = 3.19
MM1 pKa = 7.59PPAGATDD8 pKa = 3.51SFVYY12 pKa = 10.13TITDD16 pKa = 3.32GDD18 pKa = 4.4GDD20 pKa = 4.04TSTTTLTISLTDD32 pKa = 3.29SGLAASNEE40 pKa = 4.06DD41 pKa = 3.37ASVDD45 pKa = 3.65EE46 pKa = 4.54AALGIGSNPSSTAEE60 pKa = 4.18TVSGSLTDD68 pKa = 3.71NASGGAGGYY77 pKa = 8.45TYY79 pKa = 11.42ALVGSATGSHH89 pKa = 5.41GTITIDD95 pKa = 4.22PSGSWSYY102 pKa = 10.1TLTSPVDD109 pKa = 3.46GATLDD114 pKa = 3.67NGVTTEE120 pKa = 4.07NNLEE124 pKa = 4.11SFTYY128 pKa = 10.19QVTDD132 pKa = 3.57ANGNTTTSTITIDD145 pKa = 4.24VIDD148 pKa = 4.92DD149 pKa = 3.77VPTARR154 pKa = 11.84ADD156 pKa = 3.44TDD158 pKa = 4.12SVVEE162 pKa = 4.23GASTDD167 pKa = 3.66GNVLSGTGTTSGLVDD182 pKa = 3.27VLGADD187 pKa = 4.26GAAPGGAVTGVATGTDD203 pKa = 3.06TSAPVTGNLGGAGIAGTYY221 pKa = 7.35GTLILNADD229 pKa = 3.47GSYY232 pKa = 10.12TYY234 pKa = 11.23NSTANSISGNATDD247 pKa = 3.85SFVYY251 pKa = 10.1TITDD255 pKa = 3.32GDD257 pKa = 4.4GDD259 pKa = 4.04TSTTTLTISLTDD271 pKa = 3.38VSLVGDD277 pKa = 4.18DD278 pKa = 4.11EE279 pKa = 5.01SVTVYY284 pKa = 10.67EE285 pKa = 4.63KK286 pKa = 11.15ALDD289 pKa = 3.92TAQTGADD296 pKa = 3.48VAAGSITGSMPSDD309 pKa = 3.77PGEE312 pKa = 4.14TASGAVSVTGATGYY326 pKa = 10.47SIAGGSVSGNEE337 pKa = 3.95TTVVGTYY344 pKa = 7.73GTLVLNNSTGAFTYY358 pKa = 10.21TLTSPVTTTPSANDD372 pKa = 3.73GVTTALGAEE381 pKa = 4.4SFTYY385 pKa = 9.15TASDD389 pKa = 3.46ANGNTTTGTISIDD402 pKa = 3.74VVDD405 pKa = 4.38DD406 pKa = 3.45TPYY409 pKa = 11.28AVVPEE414 pKa = 5.18AIQVTNGAAPVSAPADD430 pKa = 3.73LDD432 pKa = 3.82ADD434 pKa = 3.57MSVIDD439 pKa = 4.68NYY441 pKa = 11.23GADD444 pKa = 3.41GAGTVRR450 pKa = 11.84FAPSLDD456 pKa = 3.71GADD459 pKa = 3.99SGLTHH464 pKa = 6.73NFVSITYY471 pKa = 8.28TLVSDD476 pKa = 4.52TVLEE480 pKa = 4.7AYY482 pKa = 10.22AGADD486 pKa = 3.34KK487 pKa = 10.89IFTVTLDD494 pKa = 3.71PTTATYY500 pKa = 9.29TVDD503 pKa = 3.04MDD505 pKa = 3.79GTIDD509 pKa = 4.17SIQTIDD515 pKa = 4.01FNAGGYY521 pKa = 10.3NFVGGNNSWSGFIPVGEE538 pKa = 4.46TVGSPIDD545 pKa = 4.07NNSQDD550 pKa = 4.57LLLTPSINNADD561 pKa = 4.18DD562 pKa = 3.65GTINSTANTGGIGGGASVGSNEE584 pKa = 3.97TFRR587 pKa = 11.84VDD589 pKa = 4.03FVTDD593 pKa = 3.55LRR595 pKa = 11.84GDD597 pKa = 3.87PKK599 pKa = 10.96DD600 pKa = 3.49GAGDD604 pKa = 3.9YY605 pKa = 9.02DD606 pKa = 3.92TDD608 pKa = 3.76TNRR611 pKa = 11.84DD612 pKa = 3.16HH613 pKa = 6.91MFDD616 pKa = 2.56GHH618 pKa = 5.09YY619 pKa = 9.32TVNGAVALFKK629 pKa = 10.17STNGSTVNIAAFDD642 pKa = 4.31DD643 pKa = 4.45PDD645 pKa = 4.72GNTAVGDD652 pKa = 3.84GDD654 pKa = 3.87KK655 pKa = 11.31DD656 pKa = 4.32SITGISIVWRR666 pKa = 11.84GTQYY670 pKa = 11.24VDD672 pKa = 3.7GSGDD676 pKa = 3.93PIIIPTITATNYY688 pKa = 7.78TVNGHH693 pKa = 5.33VFTVTLLGDD702 pKa = 3.77GSVNVAGVEE711 pKa = 3.98GDD713 pKa = 3.65PGSSQVGTQIAVFTGDD729 pKa = 3.17GFNSVEE735 pKa = 4.12YY736 pKa = 8.82TWAGGDD742 pKa = 3.44TFQIGDD748 pKa = 4.39FGASTLTNDD757 pKa = 4.1PVNFTIPVEE766 pKa = 4.28VVDD769 pKa = 4.37GDD771 pKa = 4.23GDD773 pKa = 3.72VSAQSNLSITAVTTTPPVALDD794 pKa = 3.64LDD796 pKa = 4.22GDD798 pKa = 4.09GVEE801 pKa = 5.61FVGLDD806 pKa = 3.01AGVTHH811 pKa = 7.96DD812 pKa = 4.2YY813 pKa = 11.45GSGLVQTAWLSADD826 pKa = 4.26DD827 pKa = 4.93GLLAHH832 pKa = 6.82DD833 pKa = 4.71TGHH836 pKa = 6.61GLDD839 pKa = 4.6IVFTDD844 pKa = 4.2DD845 pKa = 3.49AAGAEE850 pKa = 4.18TDD852 pKa = 4.38LEE854 pKa = 4.31GLRR857 pKa = 11.84LAYY860 pKa = 10.2DD861 pKa = 3.57SNGDD865 pKa = 3.67GKK867 pKa = 9.48LTAADD872 pKa = 3.59EE873 pKa = 4.72SYY875 pKa = 11.34SEE877 pKa = 4.12FGVWQDD883 pKa = 3.25ANSNGVVDD891 pKa = 4.29AGEE894 pKa = 4.36FKK896 pKa = 10.43TLAQMGITSIEE907 pKa = 4.06LTAEE911 pKa = 4.64GPGSSQADD919 pKa = 3.1GDD921 pKa = 4.83AIVHH925 pKa = 5.46ATGEE929 pKa = 4.28YY930 pKa = 8.18TMKK933 pKa = 10.67GATYY937 pKa = 10.98ALADD941 pKa = 3.59VSFATASVEE950 pKa = 4.22SQTAARR956 pKa = 11.84TAEE959 pKa = 4.02MAAITAAAAGFMMSGAAHH977 pKa = 7.38AMPLSPIAIEE987 pKa = 4.77AIAAAMQTFEE997 pKa = 4.05THH999 pKa = 5.79LPEE1002 pKa = 4.01VTVAVPEE1009 pKa = 4.32HH1010 pKa = 6.75APATPPVTFLQDD1022 pKa = 3.15GQQAHH1027 pKa = 5.91QDD1029 pKa = 3.98SEE1031 pKa = 4.78PEE1033 pKa = 4.08SHH1035 pKa = 7.36PSQHH1039 pKa = 6.62AAEE1042 pKa = 4.28QGAGHH1047 pKa = 6.82GLGDD1051 pKa = 3.82VVQHH1055 pKa = 6.05QDD1057 pKa = 3.33VSVQPLGSGEE1067 pKa = 4.13EE1068 pKa = 4.22GHH1070 pKa = 6.28VQASAAPALFGGADD1084 pKa = 3.55SGLMQALLLAAQGEE1098 pKa = 4.52QAGGSKK1104 pKa = 10.58DD1105 pKa = 3.38QGNGTGAGQGSGHH1118 pKa = 6.27GGEE1121 pKa = 4.72HH1122 pKa = 7.14GSGNLPAVQEE1132 pKa = 4.86AIAEE1136 pKa = 4.6SAGHH1140 pKa = 6.27HH1141 pKa = 6.13AVDD1144 pKa = 4.37SLIDD1148 pKa = 3.59HH1149 pKa = 7.02FAGHH1153 pKa = 5.99QGAHH1157 pKa = 6.43GAPAIAEE1164 pKa = 4.22TMGADD1169 pKa = 3.85GLDD1172 pKa = 3.55HH1173 pKa = 6.9VLGAMLAGGGLDD1185 pKa = 3.65GLHH1188 pKa = 6.73AAGLAGGFEE1197 pKa = 4.44MAAMAAAMEE1206 pKa = 4.24AHH1208 pKa = 6.04STTVAA1213 pKa = 3.19
Molecular weight: 120.74 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G6EAJ4|G6EAJ4_9SPHN Flagellar hook-associated protein 1 OS=Novosphingobium pentaromativorans US6-1 OX=1088721 GN=flgK PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84LVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.44GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 8.11VLRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.02KK41 pKa = 10.61LSAA44 pKa = 4.03
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84LVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.44GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 8.11VLRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.02KK41 pKa = 10.61LSAA44 pKa = 4.03
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1570680 |
35 |
4258 |
300.4 |
32.61 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.561 ± 0.05 | 0.932 ± 0.011 |
5.928 ± 0.022 | 5.924 ± 0.033 |
3.605 ± 0.022 | 8.687 ± 0.036 |
2.077 ± 0.019 | 5.048 ± 0.023 |
3.14 ± 0.026 | 9.84 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.56 ± 0.016 | 2.588 ± 0.022 |
5.191 ± 0.024 | 3.204 ± 0.021 |
7.296 ± 0.041 | 5.646 ± 0.025 |
5.058 ± 0.027 | 7.063 ± 0.027 |
1.425 ± 0.015 | 2.227 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
