
Coprinellus micaceus
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Agaricomycetes; Agaricomycetidae; Agaricales; Psathyrellaceae; Coprinellus
Average proteome isoelectric point is 6.98
Get precalculated fractions of proteins
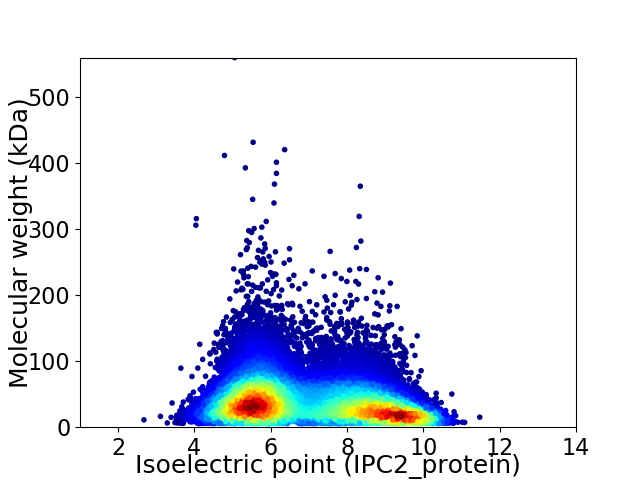
Virtual 2D-PAGE plot for 23011 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A4Y7SH21|A0A4Y7SH21_9AGAR Uncharacterized protein OS=Coprinellus micaceus OX=71717 GN=FA13DRAFT_1742332 PE=4 SV=1
MM1 pKa = 7.76GDD3 pKa = 3.42VQPVLKK9 pKa = 10.28AAYY12 pKa = 9.03PPMPNDD18 pKa = 3.89NILGPVAHH26 pKa = 7.48RR27 pKa = 11.84SLQPIPNVFEE37 pKa = 4.72LSQATTLSVDD47 pKa = 3.59DD48 pKa = 4.72GSLNGIYY55 pKa = 8.62DD56 pKa = 3.63TTTIGEE62 pKa = 4.22PSPFAPHH69 pKa = 7.49DD70 pKa = 4.39DD71 pKa = 3.8SLPPSGSIMSALVRR85 pKa = 11.84FFNLDD90 pKa = 3.23MPTNNSDD97 pKa = 3.41ATGTSSNGFDD107 pKa = 4.63SDD109 pKa = 3.41IHH111 pKa = 7.22CDD113 pKa = 4.55LIPATFDD120 pKa = 3.74SNSDD124 pKa = 3.27SSFKK128 pKa = 10.35FVPPIPTGSSSTLQQPDD145 pKa = 3.87ADD147 pKa = 3.93TGSNFDD153 pKa = 4.7SISSFDD159 pKa = 4.2FVPEE163 pKa = 4.21GNQYY167 pKa = 9.49EE168 pKa = 4.39MNMDD172 pKa = 3.19ISFPSLSNDD181 pKa = 2.68AYY183 pKa = 10.08TFGDD187 pKa = 3.26QTAYY191 pKa = 10.35SISHH195 pKa = 6.11IPFPDD200 pKa = 3.32FHH202 pKa = 7.95DD203 pKa = 4.33LAWVYY208 pKa = 10.09IASDD212 pKa = 4.09FEE214 pKa = 4.35HH215 pKa = 6.49TLNDD219 pKa = 3.43SSHH222 pKa = 6.86RR223 pKa = 11.84GLLVQAA229 pKa = 5.14
MM1 pKa = 7.76GDD3 pKa = 3.42VQPVLKK9 pKa = 10.28AAYY12 pKa = 9.03PPMPNDD18 pKa = 3.89NILGPVAHH26 pKa = 7.48RR27 pKa = 11.84SLQPIPNVFEE37 pKa = 4.72LSQATTLSVDD47 pKa = 3.59DD48 pKa = 4.72GSLNGIYY55 pKa = 8.62DD56 pKa = 3.63TTTIGEE62 pKa = 4.22PSPFAPHH69 pKa = 7.49DD70 pKa = 4.39DD71 pKa = 3.8SLPPSGSIMSALVRR85 pKa = 11.84FFNLDD90 pKa = 3.23MPTNNSDD97 pKa = 3.41ATGTSSNGFDD107 pKa = 4.63SDD109 pKa = 3.41IHH111 pKa = 7.22CDD113 pKa = 4.55LIPATFDD120 pKa = 3.74SNSDD124 pKa = 3.27SSFKK128 pKa = 10.35FVPPIPTGSSSTLQQPDD145 pKa = 3.87ADD147 pKa = 3.93TGSNFDD153 pKa = 4.7SISSFDD159 pKa = 4.2FVPEE163 pKa = 4.21GNQYY167 pKa = 9.49EE168 pKa = 4.39MNMDD172 pKa = 3.19ISFPSLSNDD181 pKa = 2.68AYY183 pKa = 10.08TFGDD187 pKa = 3.26QTAYY191 pKa = 10.35SISHH195 pKa = 6.11IPFPDD200 pKa = 3.32FHH202 pKa = 7.95DD203 pKa = 4.33LAWVYY208 pKa = 10.09IASDD212 pKa = 4.09FEE214 pKa = 4.35HH215 pKa = 6.49TLNDD219 pKa = 3.43SSHH222 pKa = 6.86RR223 pKa = 11.84GLLVQAA229 pKa = 5.14
Molecular weight: 24.71 kDa
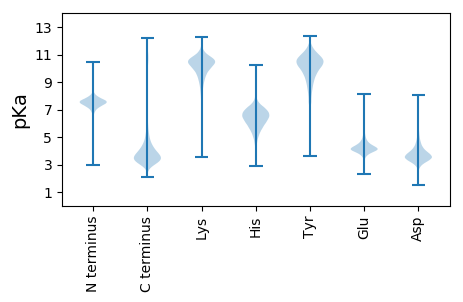
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Y7TQ73|A0A4Y7TQ73_9AGAR S-adenosyl-L-methionine-dependent methyltransferase OS=Coprinellus micaceus OX=71717 GN=FA13DRAFT_1727665 PE=4 SV=1
MM1 pKa = 7.83PFFTSRR7 pKa = 11.84RR8 pKa = 11.84TRR10 pKa = 11.84RR11 pKa = 11.84THH13 pKa = 6.4AVSTVPRR20 pKa = 11.84TSRR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84GFTLPTFGRR34 pKa = 11.84KK35 pKa = 8.59NKK37 pKa = 9.37NRR39 pKa = 11.84VAGGHH44 pKa = 6.32AAALSNPHH52 pKa = 5.09TTRR55 pKa = 11.84SGRR58 pKa = 11.84GHH60 pKa = 7.54AKK62 pKa = 10.15RR63 pKa = 11.84EE64 pKa = 3.99LRR66 pKa = 11.84AMGQGNRR73 pKa = 11.84THH75 pKa = 6.39VPLMTKK81 pKa = 9.78IKK83 pKa = 9.49RR84 pKa = 11.84ALGIRR89 pKa = 11.84SSPRR93 pKa = 11.84HH94 pKa = 4.78STHH97 pKa = 5.05TTTMGSRR104 pKa = 11.84SRR106 pKa = 11.84HH107 pKa = 5.43AIVVV111 pKa = 3.6
MM1 pKa = 7.83PFFTSRR7 pKa = 11.84RR8 pKa = 11.84TRR10 pKa = 11.84RR11 pKa = 11.84THH13 pKa = 6.4AVSTVPRR20 pKa = 11.84TSRR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84GFTLPTFGRR34 pKa = 11.84KK35 pKa = 8.59NKK37 pKa = 9.37NRR39 pKa = 11.84VAGGHH44 pKa = 6.32AAALSNPHH52 pKa = 5.09TTRR55 pKa = 11.84SGRR58 pKa = 11.84GHH60 pKa = 7.54AKK62 pKa = 10.15RR63 pKa = 11.84EE64 pKa = 3.99LRR66 pKa = 11.84AMGQGNRR73 pKa = 11.84THH75 pKa = 6.39VPLMTKK81 pKa = 9.78IKK83 pKa = 9.49RR84 pKa = 11.84ALGIRR89 pKa = 11.84SSPRR93 pKa = 11.84HH94 pKa = 4.78STHH97 pKa = 5.05TTTMGSRR104 pKa = 11.84SRR106 pKa = 11.84HH107 pKa = 5.43AIVVV111 pKa = 3.6
Molecular weight: 12.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
8897202 |
49 |
5035 |
386.6 |
42.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.261 ± 0.013 | 1.411 ± 0.008 |
5.373 ± 0.012 | 5.92 ± 0.018 |
3.673 ± 0.009 | 6.825 ± 0.017 |
2.677 ± 0.008 | 4.533 ± 0.01 |
4.469 ± 0.016 | 9.314 ± 0.019 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.972 ± 0.006 | 3.288 ± 0.008 |
6.904 ± 0.02 | 3.676 ± 0.009 |
6.67 ± 0.015 | 8.669 ± 0.018 |
6.016 ± 0.01 | 6.243 ± 0.01 |
1.53 ± 0.006 | 2.577 ± 0.007 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
