
Novosphingobium sp. FSW06-99
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Novosphingobium; unclassified Novosphingobium
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins
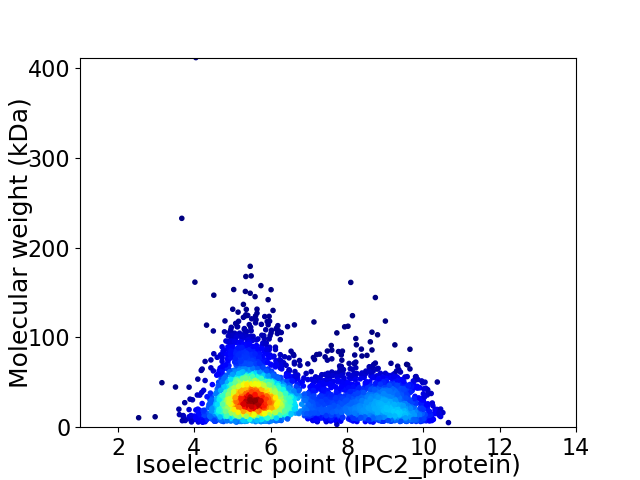
Virtual 2D-PAGE plot for 4210 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
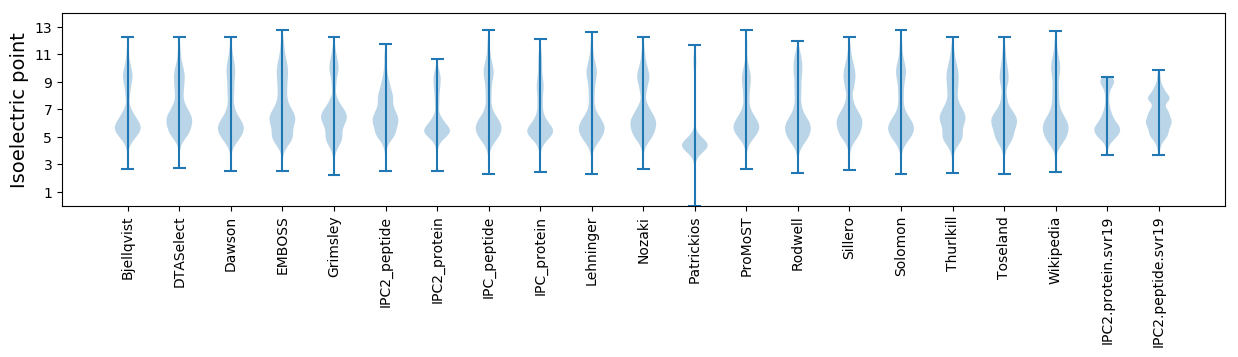
Protein with the lowest isoelectric point:
>tr|A0A124JZ66|A0A124JZ66_9SPHN Transporter OS=Novosphingobium sp. FSW06-99 OX=1739113 GN=AQZ49_05765 PE=3 SV=1
MM1 pKa = 7.57PADD4 pKa = 3.93EE5 pKa = 5.55PLDD8 pKa = 3.88CLHH11 pKa = 6.8IFSHH15 pKa = 6.16AQALFPGATIDD26 pKa = 4.52VIHH29 pKa = 6.68TPDD32 pKa = 4.12EE33 pKa = 4.74IIHH36 pKa = 6.66LDD38 pKa = 3.93VNGHH42 pKa = 6.67RR43 pKa = 11.84YY44 pKa = 7.01TFEE47 pKa = 4.31SGSDD51 pKa = 3.18DD52 pKa = 3.87DD53 pKa = 5.69EE54 pKa = 5.07YY55 pKa = 11.77VFSDD59 pKa = 3.93GEE61 pKa = 4.54TTFTIPLMDD70 pKa = 4.35FDD72 pKa = 5.29EE73 pKa = 5.18DD74 pKa = 3.92FF75 pKa = 4.57
MM1 pKa = 7.57PADD4 pKa = 3.93EE5 pKa = 5.55PLDD8 pKa = 3.88CLHH11 pKa = 6.8IFSHH15 pKa = 6.16AQALFPGATIDD26 pKa = 4.52VIHH29 pKa = 6.68TPDD32 pKa = 4.12EE33 pKa = 4.74IIHH36 pKa = 6.66LDD38 pKa = 3.93VNGHH42 pKa = 6.67RR43 pKa = 11.84YY44 pKa = 7.01TFEE47 pKa = 4.31SGSDD51 pKa = 3.18DD52 pKa = 3.87DD53 pKa = 5.69EE54 pKa = 5.07YY55 pKa = 11.77VFSDD59 pKa = 3.93GEE61 pKa = 4.54TTFTIPLMDD70 pKa = 4.35FDD72 pKa = 5.29EE73 pKa = 5.18DD74 pKa = 3.92FF75 pKa = 4.57
Molecular weight: 8.48 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A102DFR6|A0A102DFR6_9SPHN HTH lysR-type domain-containing protein OS=Novosphingobium sp. FSW06-99 OX=1739113 GN=AQZ49_00310 PE=3 SV=1
MM1 pKa = 6.36TTSDD5 pKa = 3.13RR6 pKa = 11.84RR7 pKa = 11.84TARR10 pKa = 11.84GPMRR14 pKa = 11.84AIVDD18 pKa = 3.32IGSNTVRR25 pKa = 11.84LVIYY29 pKa = 9.01GGPLRR34 pKa = 11.84APAVLHH40 pKa = 5.65NEE42 pKa = 3.87KK43 pKa = 9.17VTARR47 pKa = 11.84LGKK50 pKa = 10.2GVAEE54 pKa = 4.27NGLLGTRR61 pKa = 11.84ATTSVLAALARR72 pKa = 11.84YY73 pKa = 8.58RR74 pKa = 11.84VLLQLKK80 pKa = 8.63GVEE83 pKa = 4.19RR84 pKa = 11.84VDD86 pKa = 3.92VVATAAVRR94 pKa = 11.84DD95 pKa = 4.53AINGGAFLEE104 pKa = 4.51RR105 pKa = 11.84VAAMGFSPRR114 pKa = 11.84LLSGEE119 pKa = 4.11QEE121 pKa = 4.51AVTSAHH127 pKa = 5.98GVLGAFPGASGVVGDD142 pKa = 4.85LGGGSLEE149 pKa = 4.77LVDD152 pKa = 5.72IDD154 pKa = 5.54GDD156 pKa = 4.03TCRR159 pKa = 11.84HH160 pKa = 5.29GVSMPLGTLRR170 pKa = 11.84LGRR173 pKa = 11.84LRR175 pKa = 11.84AAGDD179 pKa = 3.36RR180 pKa = 11.84AFGQQVAKK188 pKa = 10.43ALKK191 pKa = 9.13KK192 pKa = 10.51ADD194 pKa = 3.26WAAEE198 pKa = 4.08PGSTLYY204 pKa = 11.05LVGGSLRR211 pKa = 11.84AFARR215 pKa = 11.84FAMARR220 pKa = 11.84LEE222 pKa = 4.06WPIDD226 pKa = 3.81DD227 pKa = 3.59PHH229 pKa = 8.69GFVLDD234 pKa = 3.95PADD237 pKa = 3.72GLRR240 pKa = 11.84IARR243 pKa = 11.84ALARR247 pKa = 11.84RR248 pKa = 11.84KK249 pKa = 10.1PEE251 pKa = 3.83TLRR254 pKa = 11.84PLPGISTARR263 pKa = 11.84MNSLPDD269 pKa = 3.44TAALLAVLIRR279 pKa = 11.84RR280 pKa = 11.84LQPGKK285 pKa = 10.49LVFSAWGLRR294 pKa = 11.84EE295 pKa = 3.83GLLYY299 pKa = 10.89EE300 pKa = 5.11DD301 pKa = 4.34MPAGIRR307 pKa = 11.84GQDD310 pKa = 3.58PLVAGMSAFVQEE322 pKa = 4.71HH323 pKa = 7.28GIPAQIGAMVAGWTVAAAPPSGVEE347 pKa = 3.3RR348 pKa = 11.84RR349 pKa = 11.84EE350 pKa = 3.94RR351 pKa = 11.84LRR353 pKa = 11.84LAATMLCLASAVVEE367 pKa = 4.18PNLRR371 pKa = 11.84GDD373 pKa = 3.96IARR376 pKa = 11.84DD377 pKa = 2.8WALRR381 pKa = 11.84KK382 pKa = 9.33RR383 pKa = 11.84WIGATTTEE391 pKa = 3.79RR392 pKa = 11.84VMLAVAMLGHH402 pKa = 7.65AGRR405 pKa = 11.84LDD407 pKa = 3.72VPPALAALITPAALRR422 pKa = 11.84EE423 pKa = 4.11AQTWGLATRR432 pKa = 11.84LCRR435 pKa = 11.84RR436 pKa = 11.84FSTCAPQSLTDD447 pKa = 3.48SQLTVEE453 pKa = 4.71GTRR456 pKa = 11.84LVLSVQPALAALVNEE471 pKa = 4.32GVEE474 pKa = 3.95RR475 pKa = 11.84DD476 pKa = 3.67LRR478 pKa = 11.84ALATHH483 pKa = 7.18LGLKK487 pKa = 10.5AEE489 pKa = 4.33VRR491 pKa = 3.88
MM1 pKa = 6.36TTSDD5 pKa = 3.13RR6 pKa = 11.84RR7 pKa = 11.84TARR10 pKa = 11.84GPMRR14 pKa = 11.84AIVDD18 pKa = 3.32IGSNTVRR25 pKa = 11.84LVIYY29 pKa = 9.01GGPLRR34 pKa = 11.84APAVLHH40 pKa = 5.65NEE42 pKa = 3.87KK43 pKa = 9.17VTARR47 pKa = 11.84LGKK50 pKa = 10.2GVAEE54 pKa = 4.27NGLLGTRR61 pKa = 11.84ATTSVLAALARR72 pKa = 11.84YY73 pKa = 8.58RR74 pKa = 11.84VLLQLKK80 pKa = 8.63GVEE83 pKa = 4.19RR84 pKa = 11.84VDD86 pKa = 3.92VVATAAVRR94 pKa = 11.84DD95 pKa = 4.53AINGGAFLEE104 pKa = 4.51RR105 pKa = 11.84VAAMGFSPRR114 pKa = 11.84LLSGEE119 pKa = 4.11QEE121 pKa = 4.51AVTSAHH127 pKa = 5.98GVLGAFPGASGVVGDD142 pKa = 4.85LGGGSLEE149 pKa = 4.77LVDD152 pKa = 5.72IDD154 pKa = 5.54GDD156 pKa = 4.03TCRR159 pKa = 11.84HH160 pKa = 5.29GVSMPLGTLRR170 pKa = 11.84LGRR173 pKa = 11.84LRR175 pKa = 11.84AAGDD179 pKa = 3.36RR180 pKa = 11.84AFGQQVAKK188 pKa = 10.43ALKK191 pKa = 9.13KK192 pKa = 10.51ADD194 pKa = 3.26WAAEE198 pKa = 4.08PGSTLYY204 pKa = 11.05LVGGSLRR211 pKa = 11.84AFARR215 pKa = 11.84FAMARR220 pKa = 11.84LEE222 pKa = 4.06WPIDD226 pKa = 3.81DD227 pKa = 3.59PHH229 pKa = 8.69GFVLDD234 pKa = 3.95PADD237 pKa = 3.72GLRR240 pKa = 11.84IARR243 pKa = 11.84ALARR247 pKa = 11.84RR248 pKa = 11.84KK249 pKa = 10.1PEE251 pKa = 3.83TLRR254 pKa = 11.84PLPGISTARR263 pKa = 11.84MNSLPDD269 pKa = 3.44TAALLAVLIRR279 pKa = 11.84RR280 pKa = 11.84LQPGKK285 pKa = 10.49LVFSAWGLRR294 pKa = 11.84EE295 pKa = 3.83GLLYY299 pKa = 10.89EE300 pKa = 5.11DD301 pKa = 4.34MPAGIRR307 pKa = 11.84GQDD310 pKa = 3.58PLVAGMSAFVQEE322 pKa = 4.71HH323 pKa = 7.28GIPAQIGAMVAGWTVAAAPPSGVEE347 pKa = 3.3RR348 pKa = 11.84RR349 pKa = 11.84EE350 pKa = 3.94RR351 pKa = 11.84LRR353 pKa = 11.84LAATMLCLASAVVEE367 pKa = 4.18PNLRR371 pKa = 11.84GDD373 pKa = 3.96IARR376 pKa = 11.84DD377 pKa = 2.8WALRR381 pKa = 11.84KK382 pKa = 9.33RR383 pKa = 11.84WIGATTTEE391 pKa = 3.79RR392 pKa = 11.84VMLAVAMLGHH402 pKa = 7.65AGRR405 pKa = 11.84LDD407 pKa = 3.72VPPALAALITPAALRR422 pKa = 11.84EE423 pKa = 4.11AQTWGLATRR432 pKa = 11.84LCRR435 pKa = 11.84RR436 pKa = 11.84FSTCAPQSLTDD447 pKa = 3.48SQLTVEE453 pKa = 4.71GTRR456 pKa = 11.84LVLSVQPALAALVNEE471 pKa = 4.32GVEE474 pKa = 3.95RR475 pKa = 11.84DD476 pKa = 3.67LRR478 pKa = 11.84ALATHH483 pKa = 7.18LGLKK487 pKa = 10.5AEE489 pKa = 4.33VRR491 pKa = 3.88
Molecular weight: 51.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1418537 |
29 |
4323 |
336.9 |
36.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.185 ± 0.062 | 0.854 ± 0.012 |
6.011 ± 0.03 | 4.372 ± 0.035 |
3.497 ± 0.021 | 9.032 ± 0.049 |
2.402 ± 0.02 | 4.899 ± 0.024 |
2.26 ± 0.022 | 10.042 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.277 ± 0.018 | 2.513 ± 0.039 |
5.549 ± 0.029 | 3.225 ± 0.023 |
7.125 ± 0.046 | 4.834 ± 0.038 |
5.664 ± 0.056 | 7.482 ± 0.03 |
1.519 ± 0.019 | 2.258 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
