
Streptomyces xiamenensis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces
Average proteome isoelectric point is 6.38
Get precalculated fractions of proteins
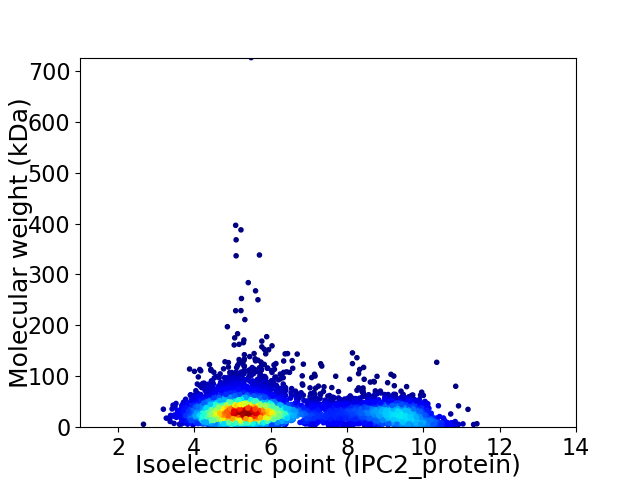
Virtual 2D-PAGE plot for 5490 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0F7FZY1|A0A0F7FZY1_9ACTN Membrane protein OS=Streptomyces xiamenensis OX=408015 GN=SXIM_50610 PE=4 SV=1
MM1 pKa = 7.76DD2 pKa = 4.04TMSNTTVRR10 pKa = 11.84RR11 pKa = 11.84LVRR14 pKa = 11.84TTAVVGALALGVTACGSSDD33 pKa = 3.92DD34 pKa = 4.16GDD36 pKa = 4.04GAGGGGGASSGLTPADD52 pKa = 3.12VEE54 pKa = 4.26AALEE58 pKa = 4.22EE59 pKa = 4.85GGSLTVWSWEE69 pKa = 3.85ATLPAAAQAFMDD81 pKa = 3.49EE82 pKa = 4.41HH83 pKa = 6.91PNVKK87 pKa = 10.26VNVVNAGTNKK97 pKa = 10.22DD98 pKa = 3.33QYY100 pKa = 10.65TALQNAISAGSGVPDD115 pKa = 3.65VAQIEE120 pKa = 4.97YY121 pKa = 10.29YY122 pKa = 11.1ALGQFTLSGALTEE135 pKa = 4.74LGSLGADD142 pKa = 3.03QLEE145 pKa = 5.14GTFSPGPWSAVQSGDD160 pKa = 3.89GIYY163 pKa = 10.74ALPTDD168 pKa = 4.68SGPMALFYY176 pKa = 11.15NKK178 pKa = 9.09TVFDD182 pKa = 3.54EE183 pKa = 4.7HH184 pKa = 8.34GIEE187 pKa = 5.82VPTTWEE193 pKa = 4.88GYY195 pKa = 10.31LEE197 pKa = 4.26AARR200 pKa = 11.84ALHH203 pKa = 6.23EE204 pKa = 5.2ADD206 pKa = 3.3PDD208 pKa = 3.6IYY210 pKa = 10.1ITNDD214 pKa = 3.0TGEE217 pKa = 4.51GGAATSMIWQAGGRR231 pKa = 11.84PYY233 pKa = 10.58TVDD236 pKa = 3.26GTDD239 pKa = 3.21VTIDD243 pKa = 3.72FSDD246 pKa = 3.79EE247 pKa = 3.65GTTAYY252 pKa = 9.53TDD254 pKa = 3.36LWQQLLDD261 pKa = 4.16EE262 pKa = 5.52DD263 pKa = 4.55LLAPITSWTDD273 pKa = 2.51EE274 pKa = 4.26WYY276 pKa = 10.5QGLGDD281 pKa = 4.26GSLATLATGAWMPPNFEE298 pKa = 4.26SGVPNAAGDD307 pKa = 3.54WRR309 pKa = 11.84AAPLPQWEE317 pKa = 4.25EE318 pKa = 3.98GANVSAEE325 pKa = 4.1NGGSSMAIPSAGKK338 pKa = 9.08NQALAYY344 pKa = 10.57AFVEE348 pKa = 4.2YY349 pKa = 10.83LNVGDD354 pKa = 4.05GVKK357 pKa = 10.35VRR359 pKa = 11.84TDD361 pKa = 2.95AGAFPATTADD371 pKa = 3.8LNSEE375 pKa = 4.16EE376 pKa = 4.22FLNTEE381 pKa = 4.21FPYY384 pKa = 10.75FGGQKK389 pKa = 10.18ANEE392 pKa = 4.06IFAEE396 pKa = 4.36SAANVGTGWSYY407 pKa = 11.68LPYY410 pKa = 10.01QVYY413 pKa = 10.9ANSIFNDD420 pKa = 3.8TAGQAYY426 pKa = 9.79ISGTTLADD434 pKa = 4.1GLKK437 pKa = 9.45AWQDD441 pKa = 3.01ASIAYY446 pKa = 9.8GKK448 pKa = 9.73EE449 pKa = 3.49QGFTVNDD456 pKa = 3.62
MM1 pKa = 7.76DD2 pKa = 4.04TMSNTTVRR10 pKa = 11.84RR11 pKa = 11.84LVRR14 pKa = 11.84TTAVVGALALGVTACGSSDD33 pKa = 3.92DD34 pKa = 4.16GDD36 pKa = 4.04GAGGGGGASSGLTPADD52 pKa = 3.12VEE54 pKa = 4.26AALEE58 pKa = 4.22EE59 pKa = 4.85GGSLTVWSWEE69 pKa = 3.85ATLPAAAQAFMDD81 pKa = 3.49EE82 pKa = 4.41HH83 pKa = 6.91PNVKK87 pKa = 10.26VNVVNAGTNKK97 pKa = 10.22DD98 pKa = 3.33QYY100 pKa = 10.65TALQNAISAGSGVPDD115 pKa = 3.65VAQIEE120 pKa = 4.97YY121 pKa = 10.29YY122 pKa = 11.1ALGQFTLSGALTEE135 pKa = 4.74LGSLGADD142 pKa = 3.03QLEE145 pKa = 5.14GTFSPGPWSAVQSGDD160 pKa = 3.89GIYY163 pKa = 10.74ALPTDD168 pKa = 4.68SGPMALFYY176 pKa = 11.15NKK178 pKa = 9.09TVFDD182 pKa = 3.54EE183 pKa = 4.7HH184 pKa = 8.34GIEE187 pKa = 5.82VPTTWEE193 pKa = 4.88GYY195 pKa = 10.31LEE197 pKa = 4.26AARR200 pKa = 11.84ALHH203 pKa = 6.23EE204 pKa = 5.2ADD206 pKa = 3.3PDD208 pKa = 3.6IYY210 pKa = 10.1ITNDD214 pKa = 3.0TGEE217 pKa = 4.51GGAATSMIWQAGGRR231 pKa = 11.84PYY233 pKa = 10.58TVDD236 pKa = 3.26GTDD239 pKa = 3.21VTIDD243 pKa = 3.72FSDD246 pKa = 3.79EE247 pKa = 3.65GTTAYY252 pKa = 9.53TDD254 pKa = 3.36LWQQLLDD261 pKa = 4.16EE262 pKa = 5.52DD263 pKa = 4.55LLAPITSWTDD273 pKa = 2.51EE274 pKa = 4.26WYY276 pKa = 10.5QGLGDD281 pKa = 4.26GSLATLATGAWMPPNFEE298 pKa = 4.26SGVPNAAGDD307 pKa = 3.54WRR309 pKa = 11.84AAPLPQWEE317 pKa = 4.25EE318 pKa = 3.98GANVSAEE325 pKa = 4.1NGGSSMAIPSAGKK338 pKa = 9.08NQALAYY344 pKa = 10.57AFVEE348 pKa = 4.2YY349 pKa = 10.83LNVGDD354 pKa = 4.05GVKK357 pKa = 10.35VRR359 pKa = 11.84TDD361 pKa = 2.95AGAFPATTADD371 pKa = 3.8LNSEE375 pKa = 4.16EE376 pKa = 4.22FLNTEE381 pKa = 4.21FPYY384 pKa = 10.75FGGQKK389 pKa = 10.18ANEE392 pKa = 4.06IFAEE396 pKa = 4.36SAANVGTGWSYY407 pKa = 11.68LPYY410 pKa = 10.01QVYY413 pKa = 10.9ANSIFNDD420 pKa = 3.8TAGQAYY426 pKa = 9.79ISGTTLADD434 pKa = 4.1GLKK437 pKa = 9.45AWQDD441 pKa = 3.01ASIAYY446 pKa = 9.8GKK448 pKa = 9.73EE449 pKa = 3.49QGFTVNDD456 pKa = 3.62
Molecular weight: 47.61 kDa
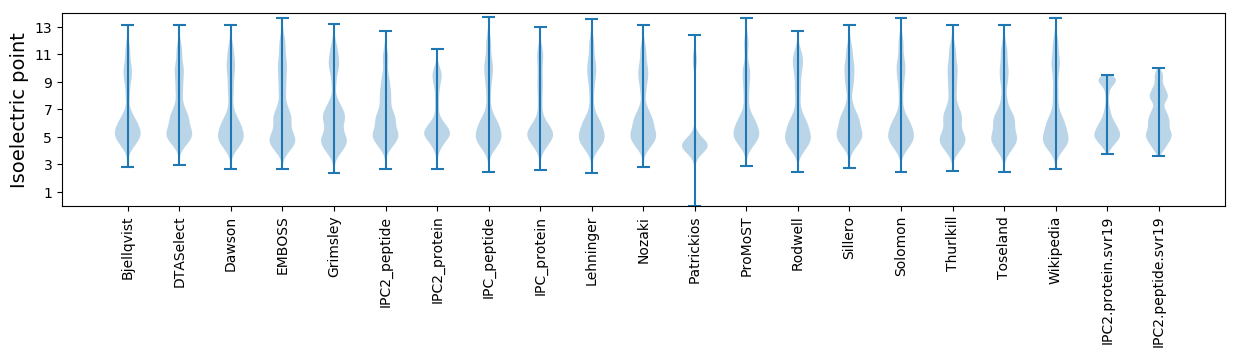
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0F7CP64|A0A0F7CP64_9ACTN Metal transporter ATPase OS=Streptomyces xiamenensis OX=408015 GN=SXIM_27320 PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AIITARR35 pKa = 11.84RR36 pKa = 11.84TKK38 pKa = 10.43GRR40 pKa = 11.84ARR42 pKa = 11.84LSAA45 pKa = 3.91
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AIITARR35 pKa = 11.84RR36 pKa = 11.84TKK38 pKa = 10.43GRR40 pKa = 11.84ARR42 pKa = 11.84LSAA45 pKa = 3.91
Molecular weight: 5.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1750436 |
37 |
6748 |
318.8 |
34.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.759 ± 0.056 | 0.743 ± 0.009 |
5.825 ± 0.028 | 6.006 ± 0.032 |
2.656 ± 0.018 | 9.754 ± 0.031 |
2.341 ± 0.018 | 3.327 ± 0.024 |
1.589 ± 0.021 | 10.588 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.719 ± 0.013 | 1.692 ± 0.023 |
6.372 ± 0.032 | 2.719 ± 0.022 |
8.326 ± 0.044 | 4.816 ± 0.023 |
6.216 ± 0.028 | 8.023 ± 0.032 |
1.504 ± 0.015 | 2.025 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
