
Parry s Lagoon virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Orbivirus; unclassified Orbivirus
Average proteome isoelectric point is 6.86
Get precalculated fractions of proteins
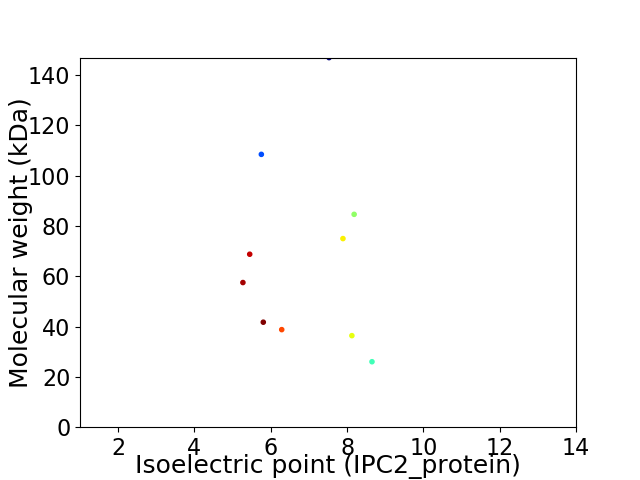
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A173G2X3|A0A173G2X3_9REOV RNA-directed RNA polymerase OS=Parry's Lagoon virus OX=1853202 PE=3 SV=1
MM1 pKa = 7.55GKK3 pKa = 8.06ITNALSKK10 pKa = 10.89FGGAVSKK17 pKa = 10.63AVKK20 pKa = 10.57SNTAKK25 pKa = 10.79KK26 pKa = 9.73IFKK29 pKa = 10.14AAIDD33 pKa = 3.79GAGRR37 pKa = 11.84VAQSEE42 pKa = 4.23LGQRR46 pKa = 11.84AISGLIEE53 pKa = 4.32GAATAALTDD62 pKa = 3.73GSYY65 pKa = 11.43GEE67 pKa = 4.21EE68 pKa = 3.78IKK70 pKa = 10.82RR71 pKa = 11.84AVILNIAGVNDD82 pKa = 3.94VVTDD86 pKa = 3.78PLNPIEE92 pKa = 4.77HH93 pKa = 7.1ALAQKK98 pKa = 10.27VNQIDD103 pKa = 3.18KK104 pKa = 10.8VMRR107 pKa = 11.84SNADD111 pKa = 2.48IDD113 pKa = 4.09KK114 pKa = 10.36YY115 pKa = 11.31GKK117 pKa = 8.83VLKK120 pKa = 10.55RR121 pKa = 11.84IDD123 pKa = 4.21GIGAEE128 pKa = 4.12LTKK131 pKa = 10.56VEE133 pKa = 4.68KK134 pKa = 10.61YY135 pKa = 10.63LQISHH140 pKa = 6.67DD141 pKa = 3.99TEE143 pKa = 3.75ISEE146 pKa = 4.24QEE148 pKa = 3.94EE149 pKa = 4.34IEE151 pKa = 4.13ALEE154 pKa = 4.28AAMKK158 pKa = 10.6AMGMVVGEE166 pKa = 4.28EE167 pKa = 4.09KK168 pKa = 10.78KK169 pKa = 10.81NLAILEE175 pKa = 4.15KK176 pKa = 10.52ALRR179 pKa = 11.84KK180 pKa = 8.56EE181 pKa = 3.87DD182 pKa = 3.37RR183 pKa = 11.84MRR185 pKa = 11.84SADD188 pKa = 3.98EE189 pKa = 3.81KK190 pKa = 11.53RR191 pKa = 11.84MIEE194 pKa = 4.04YY195 pKa = 8.41MKK197 pKa = 10.63RR198 pKa = 11.84NYY200 pKa = 9.47EE201 pKa = 4.03HH202 pKa = 7.12LAEE205 pKa = 4.45IANKK209 pKa = 9.05EE210 pKa = 3.93KK211 pKa = 10.92EE212 pKa = 4.42SIIEE216 pKa = 3.89EE217 pKa = 4.13ALEE220 pKa = 3.81QTIDD224 pKa = 3.14IGGEE228 pKa = 3.84IAEE231 pKa = 4.38HH232 pKa = 6.41LAAEE236 pKa = 4.41VPLVGEE242 pKa = 4.71GVAAGMATARR252 pKa = 11.84GAVQIYY258 pKa = 10.48KK259 pKa = 9.93LGKK262 pKa = 10.0VISDD266 pKa = 3.75LTNTPIHH273 pKa = 6.2HH274 pKa = 6.82VEE276 pKa = 4.29LPMITPEE283 pKa = 4.12GLQVLYY289 pKa = 10.54EE290 pKa = 4.26EE291 pKa = 5.23PNPTEE296 pKa = 4.18DD297 pKa = 3.65QNLLRR302 pKa = 11.84IVSSKK307 pKa = 10.24LKK309 pKa = 10.52HH310 pKa = 6.19VEE312 pKa = 4.0EE313 pKa = 4.12VDD315 pKa = 3.55KK316 pKa = 11.26EE317 pKa = 4.5VVHH320 pKa = 7.28LSEE323 pKa = 4.24KK324 pKa = 10.18VVPAVVKK331 pKa = 10.21QAATDD336 pKa = 3.72SLEE339 pKa = 4.44LGGTGKK345 pKa = 10.39GVPMRR350 pKa = 11.84TRR352 pKa = 11.84ASNHH356 pKa = 4.21VPRR359 pKa = 11.84NQRR362 pKa = 11.84PAIHH366 pKa = 7.14FYY368 pKa = 8.81TAPWDD373 pKa = 3.55SDD375 pKa = 3.5FVIILHH381 pKa = 6.25VIAPYY386 pKa = 10.71SSDD389 pKa = 3.1ASFVLALDD397 pKa = 4.41LATDD401 pKa = 3.61YY402 pKa = 11.35VGYY405 pKa = 10.69YY406 pKa = 10.18DD407 pKa = 3.75IYY409 pKa = 10.7KK410 pKa = 10.27DD411 pKa = 3.99YY412 pKa = 11.57VSDD415 pKa = 4.06CPGVDD420 pKa = 3.1SYY422 pKa = 12.18FNLQHH427 pKa = 6.82AVDD430 pKa = 4.71DD431 pKa = 4.33FLIEE435 pKa = 3.97ASAVGGSTEE444 pKa = 3.46IHH446 pKa = 6.68AEE448 pKa = 3.79RR449 pKa = 11.84LQRR452 pKa = 11.84GVGTSAIYY460 pKa = 10.5VGSQEE465 pKa = 3.85YY466 pKa = 9.56RR467 pKa = 11.84VSFEE471 pKa = 4.54AMRR474 pKa = 11.84EE475 pKa = 3.81HH476 pKa = 7.69AKK478 pKa = 10.57RR479 pKa = 11.84IVQDD483 pKa = 3.6SSIQMHH489 pKa = 6.92LLRR492 pKa = 11.84GPLSMQRR499 pKa = 11.84TSLLNALMHH508 pKa = 6.61GVTILNDD515 pKa = 3.62TPTGGMTTQCTCC527 pKa = 4.01
MM1 pKa = 7.55GKK3 pKa = 8.06ITNALSKK10 pKa = 10.89FGGAVSKK17 pKa = 10.63AVKK20 pKa = 10.57SNTAKK25 pKa = 10.79KK26 pKa = 9.73IFKK29 pKa = 10.14AAIDD33 pKa = 3.79GAGRR37 pKa = 11.84VAQSEE42 pKa = 4.23LGQRR46 pKa = 11.84AISGLIEE53 pKa = 4.32GAATAALTDD62 pKa = 3.73GSYY65 pKa = 11.43GEE67 pKa = 4.21EE68 pKa = 3.78IKK70 pKa = 10.82RR71 pKa = 11.84AVILNIAGVNDD82 pKa = 3.94VVTDD86 pKa = 3.78PLNPIEE92 pKa = 4.77HH93 pKa = 7.1ALAQKK98 pKa = 10.27VNQIDD103 pKa = 3.18KK104 pKa = 10.8VMRR107 pKa = 11.84SNADD111 pKa = 2.48IDD113 pKa = 4.09KK114 pKa = 10.36YY115 pKa = 11.31GKK117 pKa = 8.83VLKK120 pKa = 10.55RR121 pKa = 11.84IDD123 pKa = 4.21GIGAEE128 pKa = 4.12LTKK131 pKa = 10.56VEE133 pKa = 4.68KK134 pKa = 10.61YY135 pKa = 10.63LQISHH140 pKa = 6.67DD141 pKa = 3.99TEE143 pKa = 3.75ISEE146 pKa = 4.24QEE148 pKa = 3.94EE149 pKa = 4.34IEE151 pKa = 4.13ALEE154 pKa = 4.28AAMKK158 pKa = 10.6AMGMVVGEE166 pKa = 4.28EE167 pKa = 4.09KK168 pKa = 10.78KK169 pKa = 10.81NLAILEE175 pKa = 4.15KK176 pKa = 10.52ALRR179 pKa = 11.84KK180 pKa = 8.56EE181 pKa = 3.87DD182 pKa = 3.37RR183 pKa = 11.84MRR185 pKa = 11.84SADD188 pKa = 3.98EE189 pKa = 3.81KK190 pKa = 11.53RR191 pKa = 11.84MIEE194 pKa = 4.04YY195 pKa = 8.41MKK197 pKa = 10.63RR198 pKa = 11.84NYY200 pKa = 9.47EE201 pKa = 4.03HH202 pKa = 7.12LAEE205 pKa = 4.45IANKK209 pKa = 9.05EE210 pKa = 3.93KK211 pKa = 10.92EE212 pKa = 4.42SIIEE216 pKa = 3.89EE217 pKa = 4.13ALEE220 pKa = 3.81QTIDD224 pKa = 3.14IGGEE228 pKa = 3.84IAEE231 pKa = 4.38HH232 pKa = 6.41LAAEE236 pKa = 4.41VPLVGEE242 pKa = 4.71GVAAGMATARR252 pKa = 11.84GAVQIYY258 pKa = 10.48KK259 pKa = 9.93LGKK262 pKa = 10.0VISDD266 pKa = 3.75LTNTPIHH273 pKa = 6.2HH274 pKa = 6.82VEE276 pKa = 4.29LPMITPEE283 pKa = 4.12GLQVLYY289 pKa = 10.54EE290 pKa = 4.26EE291 pKa = 5.23PNPTEE296 pKa = 4.18DD297 pKa = 3.65QNLLRR302 pKa = 11.84IVSSKK307 pKa = 10.24LKK309 pKa = 10.52HH310 pKa = 6.19VEE312 pKa = 4.0EE313 pKa = 4.12VDD315 pKa = 3.55KK316 pKa = 11.26EE317 pKa = 4.5VVHH320 pKa = 7.28LSEE323 pKa = 4.24KK324 pKa = 10.18VVPAVVKK331 pKa = 10.21QAATDD336 pKa = 3.72SLEE339 pKa = 4.44LGGTGKK345 pKa = 10.39GVPMRR350 pKa = 11.84TRR352 pKa = 11.84ASNHH356 pKa = 4.21VPRR359 pKa = 11.84NQRR362 pKa = 11.84PAIHH366 pKa = 7.14FYY368 pKa = 8.81TAPWDD373 pKa = 3.55SDD375 pKa = 3.5FVIILHH381 pKa = 6.25VIAPYY386 pKa = 10.71SSDD389 pKa = 3.1ASFVLALDD397 pKa = 4.41LATDD401 pKa = 3.61YY402 pKa = 11.35VGYY405 pKa = 10.69YY406 pKa = 10.18DD407 pKa = 3.75IYY409 pKa = 10.7KK410 pKa = 10.27DD411 pKa = 3.99YY412 pKa = 11.57VSDD415 pKa = 4.06CPGVDD420 pKa = 3.1SYY422 pKa = 12.18FNLQHH427 pKa = 6.82AVDD430 pKa = 4.71DD431 pKa = 4.33FLIEE435 pKa = 3.97ASAVGGSTEE444 pKa = 3.46IHH446 pKa = 6.68AEE448 pKa = 3.79RR449 pKa = 11.84LQRR452 pKa = 11.84GVGTSAIYY460 pKa = 10.5VGSQEE465 pKa = 3.85YY466 pKa = 9.56RR467 pKa = 11.84VSFEE471 pKa = 4.54AMRR474 pKa = 11.84EE475 pKa = 3.81HH476 pKa = 7.69AKK478 pKa = 10.57RR479 pKa = 11.84IVQDD483 pKa = 3.6SSIQMHH489 pKa = 6.92LLRR492 pKa = 11.84GPLSMQRR499 pKa = 11.84TSLLNALMHH508 pKa = 6.61GVTILNDD515 pKa = 3.62TPTGGMTTQCTCC527 pKa = 4.01
Molecular weight: 57.53 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A173G2X7|A0A173G2X7_9REOV Non-structural protein NS3 OS=Parry's Lagoon virus OX=1853202 PE=3 SV=1
MM1 pKa = 7.47LALADD6 pKa = 3.97SLVGKK11 pKa = 9.99GPKK14 pKa = 9.16QDD16 pKa = 3.29VVVTVVPPEE25 pKa = 3.94KK26 pKa = 10.4FGLLKK31 pKa = 10.19DD32 pKa = 3.74TSNEE36 pKa = 3.53EE37 pKa = 3.82RR38 pKa = 11.84DD39 pKa = 3.29RR40 pKa = 11.84DD41 pKa = 3.66GALSVLTNAIVSATGTNEE59 pKa = 3.79TQKK62 pKa = 10.88NEE64 pKa = 3.83KK65 pKa = 9.02ATFGAISEE73 pKa = 4.28ALKK76 pKa = 10.72DD77 pKa = 3.86DD78 pKa = 3.55NVTRR82 pKa = 11.84RR83 pKa = 11.84SKK85 pKa = 10.69KK86 pKa = 8.29IAYY89 pKa = 8.19SKK91 pKa = 11.08SVFEE95 pKa = 4.37LEE97 pKa = 3.61KK98 pKa = 10.67DD99 pKa = 2.72IRR101 pKa = 11.84RR102 pKa = 11.84YY103 pKa = 10.59RR104 pKa = 11.84NTIRR108 pKa = 11.84LLEE111 pKa = 4.2CARR114 pKa = 11.84VGCAIVVLLFSAALSIMDD132 pKa = 4.72FLSKK136 pKa = 10.91DD137 pKa = 4.14VITKK141 pKa = 8.16MATFVGDD148 pKa = 3.33HH149 pKa = 6.58GSADD153 pKa = 3.55FAAMTRR159 pKa = 11.84LRR161 pKa = 11.84AIRR164 pKa = 11.84GAVNMALITCGTAVLITGRR183 pKa = 11.84NVAIMKK189 pKa = 8.81QKK191 pKa = 10.16KK192 pKa = 9.41RR193 pKa = 11.84GMKK196 pKa = 9.57RR197 pKa = 11.84DD198 pKa = 3.39LVKK201 pKa = 10.8RR202 pKa = 11.84KK203 pKa = 9.8AYY205 pKa = 10.55LGAIDD210 pKa = 5.9DD211 pKa = 4.5MNLLTPYY218 pKa = 9.68AIEE221 pKa = 4.43MDD223 pKa = 4.08PPTYY227 pKa = 10.54AAAKK231 pKa = 10.12APYY234 pKa = 9.75EE235 pKa = 3.99YY236 pKa = 11.06DD237 pKa = 3.18LL238 pKa = 5.4
MM1 pKa = 7.47LALADD6 pKa = 3.97SLVGKK11 pKa = 9.99GPKK14 pKa = 9.16QDD16 pKa = 3.29VVVTVVPPEE25 pKa = 3.94KK26 pKa = 10.4FGLLKK31 pKa = 10.19DD32 pKa = 3.74TSNEE36 pKa = 3.53EE37 pKa = 3.82RR38 pKa = 11.84DD39 pKa = 3.29RR40 pKa = 11.84DD41 pKa = 3.66GALSVLTNAIVSATGTNEE59 pKa = 3.79TQKK62 pKa = 10.88NEE64 pKa = 3.83KK65 pKa = 9.02ATFGAISEE73 pKa = 4.28ALKK76 pKa = 10.72DD77 pKa = 3.86DD78 pKa = 3.55NVTRR82 pKa = 11.84RR83 pKa = 11.84SKK85 pKa = 10.69KK86 pKa = 8.29IAYY89 pKa = 8.19SKK91 pKa = 11.08SVFEE95 pKa = 4.37LEE97 pKa = 3.61KK98 pKa = 10.67DD99 pKa = 2.72IRR101 pKa = 11.84RR102 pKa = 11.84YY103 pKa = 10.59RR104 pKa = 11.84NTIRR108 pKa = 11.84LLEE111 pKa = 4.2CARR114 pKa = 11.84VGCAIVVLLFSAALSIMDD132 pKa = 4.72FLSKK136 pKa = 10.91DD137 pKa = 4.14VITKK141 pKa = 8.16MATFVGDD148 pKa = 3.33HH149 pKa = 6.58GSADD153 pKa = 3.55FAAMTRR159 pKa = 11.84LRR161 pKa = 11.84AIRR164 pKa = 11.84GAVNMALITCGTAVLITGRR183 pKa = 11.84NVAIMKK189 pKa = 8.81QKK191 pKa = 10.16KK192 pKa = 9.41RR193 pKa = 11.84GMKK196 pKa = 9.57RR197 pKa = 11.84DD198 pKa = 3.39LVKK201 pKa = 10.8RR202 pKa = 11.84KK203 pKa = 9.8AYY205 pKa = 10.55LGAIDD210 pKa = 5.9DD211 pKa = 4.5MNLLTPYY218 pKa = 9.68AIEE221 pKa = 4.43MDD223 pKa = 4.08PPTYY227 pKa = 10.54AAAKK231 pKa = 10.12APYY234 pKa = 9.75EE235 pKa = 3.99YY236 pKa = 11.06DD237 pKa = 3.18LL238 pKa = 5.4
Molecular weight: 26.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6043 |
238 |
1290 |
604.3 |
68.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.314 ± 0.717 | 1.092 ± 0.205 |
6.619 ± 0.363 | 6.354 ± 0.406 |
3.376 ± 0.372 | 5.891 ± 0.391 |
2.664 ± 0.198 | 6.272 ± 0.367 |
5.891 ± 0.661 | 8.804 ± 0.389 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.161 ± 0.223 | 3.756 ± 0.307 |
4.154 ± 0.315 | 3.227 ± 0.293 |
6.851 ± 0.426 | 6.47 ± 0.446 |
5.941 ± 0.387 | 7.083 ± 0.422 |
1.191 ± 0.252 | 3.872 ± 0.295 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
