
Monaibacterium marinum
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Monaibacterium
Average proteome isoelectric point is 5.94
Get precalculated fractions of proteins
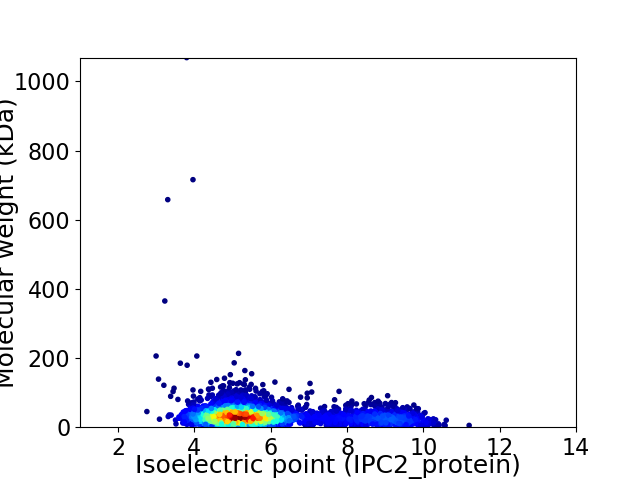
Virtual 2D-PAGE plot for 3548 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2C9CRK9|A0A2C9CRK9_9RHOB Protein translocase subunit SecA OS=Monaibacterium marinum OX=1690039 GN=secA PE=3 SV=1
MM1 pKa = 5.65QTYY4 pKa = 10.21RR5 pKa = 11.84IISMTMGMMLSAGTAMACAPGEE27 pKa = 4.13DD28 pKa = 3.88LRR30 pKa = 11.84ATLGFADD37 pKa = 4.69LTAAEE42 pKa = 4.04QDD44 pKa = 3.21GFARR48 pKa = 11.84LFVPEE53 pKa = 4.02QAVLTSEE60 pKa = 4.98NIAIDD65 pKa = 3.69DD66 pKa = 3.79AFGFARR72 pKa = 11.84CDD74 pKa = 3.41LGGMGDD80 pKa = 3.61DD81 pKa = 3.85TLITFALPGVFCEE94 pKa = 4.44EE95 pKa = 4.11NCLLWALHH103 pKa = 5.47RR104 pKa = 11.84TEE106 pKa = 4.6EE107 pKa = 4.62DD108 pKa = 2.84DD109 pKa = 3.51WQIILAAEE117 pKa = 4.39GAVTVAGSYY126 pKa = 11.1SMGWPDD132 pKa = 4.54LVAHH136 pKa = 6.6GEE138 pKa = 4.14GFADD142 pKa = 3.6IVHH145 pKa = 6.54KK146 pKa = 10.68FDD148 pKa = 3.36GASYY152 pKa = 10.21HH153 pKa = 7.45DD154 pKa = 4.31EE155 pKa = 4.47LEE157 pKa = 4.0GLIYY161 pKa = 11.0GEE163 pKa = 5.04VFDD166 pKa = 5.96LPQATSWQADD176 pKa = 3.44EE177 pKa = 4.49FDD179 pKa = 3.98FVGVVPGSPGPAGEE193 pKa = 4.16AVIALEE199 pKa = 5.23AVAQQLDD206 pKa = 3.43ISIDD210 pKa = 3.41GLAAGLTDD218 pKa = 4.89LNGDD222 pKa = 4.75EE223 pKa = 4.24IPEE226 pKa = 4.26VIVQGEE232 pKa = 4.18GAEE235 pKa = 4.17FCAADD240 pKa = 4.05GCRR243 pKa = 11.84TWIVSITAPNATILADD259 pKa = 3.54VTAQGTPEE267 pKa = 3.8IAASTNGAYY276 pKa = 9.88RR277 pKa = 11.84DD278 pKa = 3.67IIIWSNAGARR288 pKa = 11.84VLRR291 pKa = 11.84HH292 pKa = 6.79DD293 pKa = 4.16GNTYY297 pKa = 10.01HH298 pKa = 7.42
MM1 pKa = 5.65QTYY4 pKa = 10.21RR5 pKa = 11.84IISMTMGMMLSAGTAMACAPGEE27 pKa = 4.13DD28 pKa = 3.88LRR30 pKa = 11.84ATLGFADD37 pKa = 4.69LTAAEE42 pKa = 4.04QDD44 pKa = 3.21GFARR48 pKa = 11.84LFVPEE53 pKa = 4.02QAVLTSEE60 pKa = 4.98NIAIDD65 pKa = 3.69DD66 pKa = 3.79AFGFARR72 pKa = 11.84CDD74 pKa = 3.41LGGMGDD80 pKa = 3.61DD81 pKa = 3.85TLITFALPGVFCEE94 pKa = 4.44EE95 pKa = 4.11NCLLWALHH103 pKa = 5.47RR104 pKa = 11.84TEE106 pKa = 4.6EE107 pKa = 4.62DD108 pKa = 2.84DD109 pKa = 3.51WQIILAAEE117 pKa = 4.39GAVTVAGSYY126 pKa = 11.1SMGWPDD132 pKa = 4.54LVAHH136 pKa = 6.6GEE138 pKa = 4.14GFADD142 pKa = 3.6IVHH145 pKa = 6.54KK146 pKa = 10.68FDD148 pKa = 3.36GASYY152 pKa = 10.21HH153 pKa = 7.45DD154 pKa = 4.31EE155 pKa = 4.47LEE157 pKa = 4.0GLIYY161 pKa = 11.0GEE163 pKa = 5.04VFDD166 pKa = 5.96LPQATSWQADD176 pKa = 3.44EE177 pKa = 4.49FDD179 pKa = 3.98FVGVVPGSPGPAGEE193 pKa = 4.16AVIALEE199 pKa = 5.23AVAQQLDD206 pKa = 3.43ISIDD210 pKa = 3.41GLAAGLTDD218 pKa = 4.89LNGDD222 pKa = 4.75EE223 pKa = 4.24IPEE226 pKa = 4.26VIVQGEE232 pKa = 4.18GAEE235 pKa = 4.17FCAADD240 pKa = 4.05GCRR243 pKa = 11.84TWIVSITAPNATILADD259 pKa = 3.54VTAQGTPEE267 pKa = 3.8IAASTNGAYY276 pKa = 9.88RR277 pKa = 11.84DD278 pKa = 3.67IIIWSNAGARR288 pKa = 11.84VLRR291 pKa = 11.84HH292 pKa = 6.79DD293 pKa = 4.16GNTYY297 pKa = 10.01HH298 pKa = 7.42
Molecular weight: 31.51 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2C9CNC5|A0A2C9CNC5_9RHOB ADP-ribose pyrophosphatase YjhB NUDIX family OS=Monaibacterium marinum OX=1690039 GN=SAMN06273572_101565 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 8.96RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.62GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.45NGRR28 pKa = 11.84KK29 pKa = 9.03ILNARR34 pKa = 11.84RR35 pKa = 11.84AQGRR39 pKa = 11.84KK40 pKa = 9.02HH41 pKa = 6.53LSAA44 pKa = 5.78
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 8.96RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.62GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.45NGRR28 pKa = 11.84KK29 pKa = 9.03ILNARR34 pKa = 11.84RR35 pKa = 11.84AQGRR39 pKa = 11.84KK40 pKa = 9.02HH41 pKa = 6.53LSAA44 pKa = 5.78
Molecular weight: 5.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1142599 |
30 |
10629 |
322.0 |
34.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.229 ± 0.063 | 0.868 ± 0.016 |
6.347 ± 0.063 | 5.533 ± 0.05 |
3.606 ± 0.032 | 8.681 ± 0.078 |
2.035 ± 0.03 | 5.554 ± 0.035 |
2.658 ± 0.038 | 9.836 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.883 ± 0.031 | 2.798 ± 0.032 |
4.77 ± 0.056 | 3.486 ± 0.026 |
6.432 ± 0.064 | 5.398 ± 0.045 |
5.925 ± 0.072 | 7.419 ± 0.038 |
1.339 ± 0.022 | 2.206 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
