
Corynebacterium phage Lederberg
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; Samwavirus; unclassified Samwavirus
Average proteome isoelectric point is 5.77
Get precalculated fractions of proteins
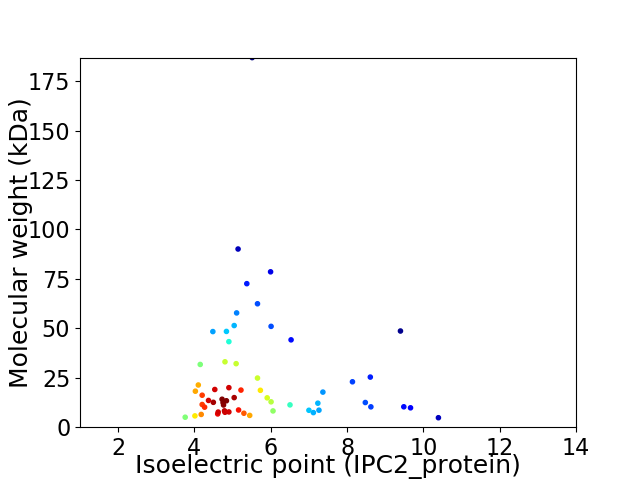
Virtual 2D-PAGE plot for 60 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
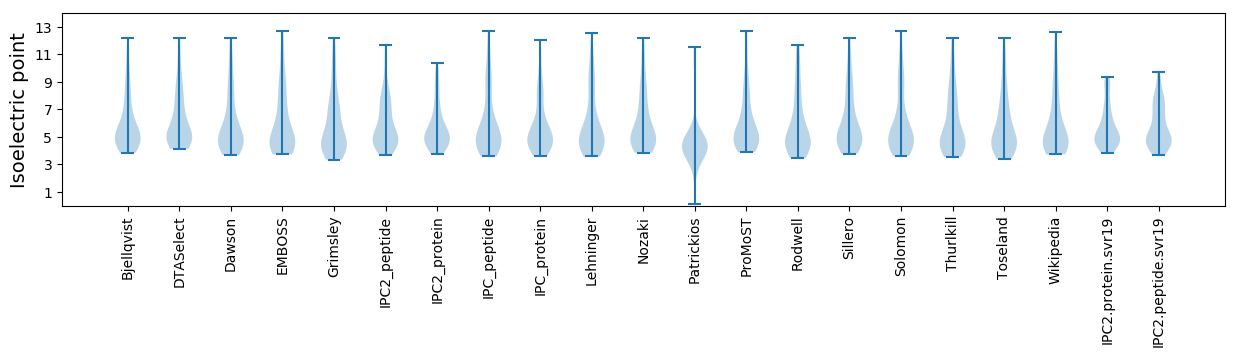
Protein with the lowest isoelectric point:
>tr|A0A4Y6EXC4|A0A4Y6EXC4_9CAUD Uncharacterized protein OS=Corynebacterium phage Lederberg OX=2588502 GN=47 PE=4 SV=1
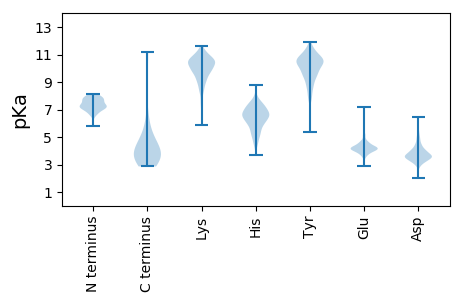
MM1 pKa = 7.88TDD3 pKa = 3.23PDD5 pKa = 4.4FPDD8 pKa = 3.66SYY10 pKa = 11.23KK11 pKa = 10.64LLHH14 pKa = 6.18GAPYY18 pKa = 10.72DD19 pKa = 4.53DD20 pKa = 6.43DD21 pKa = 5.09PGPWPYY27 pKa = 10.86LAVIAFAAIAAVAILVASYY46 pKa = 10.82LL47 pKa = 3.74
MM1 pKa = 7.88TDD3 pKa = 3.23PDD5 pKa = 4.4FPDD8 pKa = 3.66SYY10 pKa = 11.23KK11 pKa = 10.64LLHH14 pKa = 6.18GAPYY18 pKa = 10.72DD19 pKa = 4.53DD20 pKa = 6.43DD21 pKa = 5.09PGPWPYY27 pKa = 10.86LAVIAFAAIAAVAILVASYY46 pKa = 10.82LL47 pKa = 3.74
Molecular weight: 5.05 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Y6EXB5|A0A4Y6EXB5_9CAUD Uncharacterized protein OS=Corynebacterium phage Lederberg OX=2588502 GN=37 PE=4 SV=1
MM1 pKa = 8.05PEE3 pKa = 3.79TTNRR7 pKa = 11.84PTPHH11 pKa = 7.17ALDD14 pKa = 3.63SMRR17 pKa = 11.84GGAGATAPTGGSKK30 pKa = 10.93DD31 pKa = 3.81EE32 pKa = 4.21ITVSVQRR39 pKa = 11.84RR40 pKa = 11.84EE41 pKa = 3.63RR42 pKa = 11.84GGRR45 pKa = 11.84VRR47 pKa = 11.84WIGRR51 pKa = 11.84YY52 pKa = 9.18RR53 pKa = 11.84GLDD56 pKa = 3.02GKK58 pKa = 10.34EE59 pKa = 3.48RR60 pKa = 11.84SRR62 pKa = 11.84SFEE65 pKa = 3.7KK66 pKa = 10.72RR67 pKa = 11.84RR68 pKa = 11.84DD69 pKa = 3.41AVTWVAEE76 pKa = 4.44KK77 pKa = 10.75KK78 pKa = 9.81RR79 pKa = 11.84DD80 pKa = 3.58LRR82 pKa = 11.84RR83 pKa = 11.84GDD85 pKa = 3.58WIDD88 pKa = 3.39PAEE91 pKa = 4.02QKK93 pKa = 8.97ITIGEE98 pKa = 4.26LARR101 pKa = 11.84EE102 pKa = 4.07RR103 pKa = 11.84TMMARR108 pKa = 11.84TTNTKK113 pKa = 7.59ATRR116 pKa = 11.84RR117 pKa = 11.84FFEE120 pKa = 4.82ANLGPLEE127 pKa = 4.3GAAIGTVRR135 pKa = 11.84ASHH138 pKa = 5.1MRR140 pKa = 11.84EE141 pKa = 3.37WHH143 pKa = 5.06TMLRR147 pKa = 11.84NGRR150 pKa = 11.84PWQSGSLSPATIDD163 pKa = 3.34GLTVQMRR170 pKa = 11.84TLMKK174 pKa = 10.35QAVADD179 pKa = 4.09GLISRR184 pKa = 11.84SPAAGLEE191 pKa = 4.11RR192 pKa = 11.84VGRR195 pKa = 11.84DD196 pKa = 3.14VVVIPRR202 pKa = 11.84DD203 pKa = 3.26IPTPAEE209 pKa = 3.78VGALIRR215 pKa = 11.84AFEE218 pKa = 4.07TGIRR222 pKa = 11.84APEE225 pKa = 4.02PLVPQYY231 pKa = 10.97RR232 pKa = 11.84RR233 pKa = 11.84DD234 pKa = 3.42GRR236 pKa = 11.84RR237 pKa = 11.84KK238 pKa = 8.85PRR240 pKa = 11.84GWLLKK245 pKa = 8.91PAPWMARR252 pKa = 11.84AVRR255 pKa = 11.84LAIATGMRR263 pKa = 11.84PSEE266 pKa = 4.07VAGMRR271 pKa = 11.84VRR273 pKa = 11.84SLDD276 pKa = 3.44QLTGSVDD283 pKa = 3.69VVEE286 pKa = 4.36QAARR290 pKa = 11.84HH291 pKa = 4.38STEE294 pKa = 3.8TVEE297 pKa = 5.32LKK299 pKa = 10.07TEE301 pKa = 4.38ASRR304 pKa = 11.84RR305 pKa = 11.84WLPVDD310 pKa = 3.92AEE312 pKa = 4.42TLHH315 pKa = 7.18VLVDD319 pKa = 3.7WAQDD323 pKa = 3.18RR324 pKa = 11.84DD325 pKa = 3.94LGPDD329 pKa = 2.8ARR331 pKa = 11.84LFHH334 pKa = 6.45VRR336 pKa = 11.84GGNPATAEE344 pKa = 4.37AIGRR348 pKa = 11.84LMRR351 pKa = 11.84SAVDD355 pKa = 3.19AGVWRR360 pKa = 11.84GGMTFHH366 pKa = 7.62SLRR369 pKa = 11.84HH370 pKa = 4.92FHH372 pKa = 7.06ASALIAAGLDD382 pKa = 3.32VAVVSRR388 pKa = 11.84RR389 pKa = 11.84LGHH392 pKa = 6.35EE393 pKa = 3.99NMSTTLEE400 pKa = 4.57VYY402 pKa = 8.49THH404 pKa = 7.03LWPGSEE410 pKa = 3.9EE411 pKa = 4.31RR412 pKa = 11.84EE413 pKa = 4.0RR414 pKa = 11.84EE415 pKa = 3.8AATALVRR422 pKa = 11.84DD423 pKa = 3.83VCGTGGRR430 pKa = 11.84GEE432 pKa = 4.48GSDD435 pKa = 3.5SGSVQVSS442 pKa = 3.17
MM1 pKa = 8.05PEE3 pKa = 3.79TTNRR7 pKa = 11.84PTPHH11 pKa = 7.17ALDD14 pKa = 3.63SMRR17 pKa = 11.84GGAGATAPTGGSKK30 pKa = 10.93DD31 pKa = 3.81EE32 pKa = 4.21ITVSVQRR39 pKa = 11.84RR40 pKa = 11.84EE41 pKa = 3.63RR42 pKa = 11.84GGRR45 pKa = 11.84VRR47 pKa = 11.84WIGRR51 pKa = 11.84YY52 pKa = 9.18RR53 pKa = 11.84GLDD56 pKa = 3.02GKK58 pKa = 10.34EE59 pKa = 3.48RR60 pKa = 11.84SRR62 pKa = 11.84SFEE65 pKa = 3.7KK66 pKa = 10.72RR67 pKa = 11.84RR68 pKa = 11.84DD69 pKa = 3.41AVTWVAEE76 pKa = 4.44KK77 pKa = 10.75KK78 pKa = 9.81RR79 pKa = 11.84DD80 pKa = 3.58LRR82 pKa = 11.84RR83 pKa = 11.84GDD85 pKa = 3.58WIDD88 pKa = 3.39PAEE91 pKa = 4.02QKK93 pKa = 8.97ITIGEE98 pKa = 4.26LARR101 pKa = 11.84EE102 pKa = 4.07RR103 pKa = 11.84TMMARR108 pKa = 11.84TTNTKK113 pKa = 7.59ATRR116 pKa = 11.84RR117 pKa = 11.84FFEE120 pKa = 4.82ANLGPLEE127 pKa = 4.3GAAIGTVRR135 pKa = 11.84ASHH138 pKa = 5.1MRR140 pKa = 11.84EE141 pKa = 3.37WHH143 pKa = 5.06TMLRR147 pKa = 11.84NGRR150 pKa = 11.84PWQSGSLSPATIDD163 pKa = 3.34GLTVQMRR170 pKa = 11.84TLMKK174 pKa = 10.35QAVADD179 pKa = 4.09GLISRR184 pKa = 11.84SPAAGLEE191 pKa = 4.11RR192 pKa = 11.84VGRR195 pKa = 11.84DD196 pKa = 3.14VVVIPRR202 pKa = 11.84DD203 pKa = 3.26IPTPAEE209 pKa = 3.78VGALIRR215 pKa = 11.84AFEE218 pKa = 4.07TGIRR222 pKa = 11.84APEE225 pKa = 4.02PLVPQYY231 pKa = 10.97RR232 pKa = 11.84RR233 pKa = 11.84DD234 pKa = 3.42GRR236 pKa = 11.84RR237 pKa = 11.84KK238 pKa = 8.85PRR240 pKa = 11.84GWLLKK245 pKa = 8.91PAPWMARR252 pKa = 11.84AVRR255 pKa = 11.84LAIATGMRR263 pKa = 11.84PSEE266 pKa = 4.07VAGMRR271 pKa = 11.84VRR273 pKa = 11.84SLDD276 pKa = 3.44QLTGSVDD283 pKa = 3.69VVEE286 pKa = 4.36QAARR290 pKa = 11.84HH291 pKa = 4.38STEE294 pKa = 3.8TVEE297 pKa = 5.32LKK299 pKa = 10.07TEE301 pKa = 4.38ASRR304 pKa = 11.84RR305 pKa = 11.84WLPVDD310 pKa = 3.92AEE312 pKa = 4.42TLHH315 pKa = 7.18VLVDD319 pKa = 3.7WAQDD323 pKa = 3.18RR324 pKa = 11.84DD325 pKa = 3.94LGPDD329 pKa = 2.8ARR331 pKa = 11.84LFHH334 pKa = 6.45VRR336 pKa = 11.84GGNPATAEE344 pKa = 4.37AIGRR348 pKa = 11.84LMRR351 pKa = 11.84SAVDD355 pKa = 3.19AGVWRR360 pKa = 11.84GGMTFHH366 pKa = 7.62SLRR369 pKa = 11.84HH370 pKa = 4.92FHH372 pKa = 7.06ASALIAAGLDD382 pKa = 3.32VAVVSRR388 pKa = 11.84RR389 pKa = 11.84LGHH392 pKa = 6.35EE393 pKa = 3.99NMSTTLEE400 pKa = 4.57VYY402 pKa = 8.49THH404 pKa = 7.03LWPGSEE410 pKa = 3.9EE411 pKa = 4.31RR412 pKa = 11.84EE413 pKa = 4.0RR414 pKa = 11.84EE415 pKa = 3.8AATALVRR422 pKa = 11.84DD423 pKa = 3.83VCGTGGRR430 pKa = 11.84GEE432 pKa = 4.48GSDD435 pKa = 3.5SGSVQVSS442 pKa = 3.17
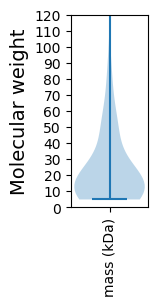
Molecular weight: 48.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
14027 |
45 |
1803 |
233.8 |
25.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.775 ± 0.663 | 0.606 ± 0.103 |
7.065 ± 0.38 | 6.138 ± 0.261 |
2.452 ± 0.194 | 8.598 ± 0.518 |
2.424 ± 0.277 | 4.869 ± 0.192 |
3.301 ± 0.228 | 7.721 ± 0.357 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.445 ± 0.112 | 2.438 ± 0.185 |
5.789 ± 0.385 | 3.45 ± 0.189 |
7.115 ± 0.467 | 4.748 ± 0.412 |
6.794 ± 0.318 | 6.88 ± 0.231 |
2.231 ± 0.15 | 2.16 ± 0.229 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
