
Flavobacterium aurantiibacter
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Flavobacterium
Average proteome isoelectric point is 6.75
Get precalculated fractions of proteins
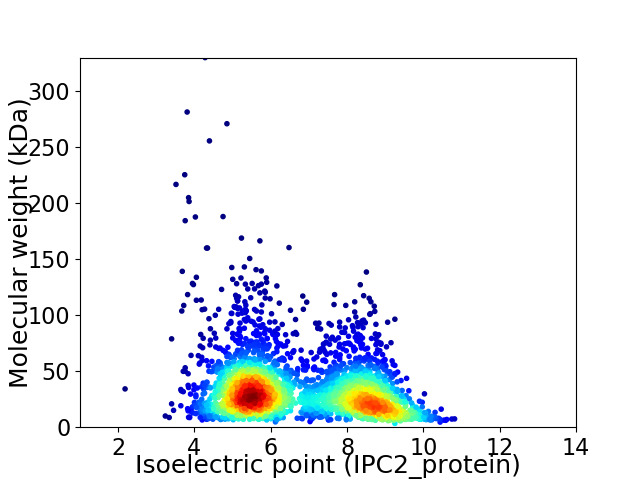
Virtual 2D-PAGE plot for 2902 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A255ZQK0|A0A255ZQK0_9FLAO Uncharacterized protein OS=Flavobacterium aurantiibacter OX=2023067 GN=CHX27_09375 PE=4 SV=1
MM1 pKa = 7.58KK2 pKa = 10.36KK3 pKa = 9.42IQLFFLALSTALTLSGCEE21 pKa = 3.99TDD23 pKa = 5.45GGDD26 pKa = 4.15SKK28 pKa = 11.84LNLIEE33 pKa = 4.66AAVPDD38 pKa = 3.56ITINDD43 pKa = 3.76TSDD46 pKa = 3.43EE47 pKa = 4.52IINLVAVTNGGDD59 pKa = 3.31INLSLTLDD67 pKa = 3.31KK68 pKa = 11.55GFGKK72 pKa = 10.27VSSMDD77 pKa = 3.25VVLFYY82 pKa = 11.11YY83 pKa = 10.58KK84 pKa = 10.38SDD86 pKa = 3.06GSVFRR91 pKa = 11.84SVLQSGITEE100 pKa = 4.6FPATVSITKK109 pKa = 10.45NSIFDD114 pKa = 3.42AFEE117 pKa = 4.24QINVPADD124 pKa = 3.59FEE126 pKa = 4.71VGDD129 pKa = 4.34EE130 pKa = 4.23LLITADD136 pKa = 3.43LTLPNGTVIKK146 pKa = 9.75MYY148 pKa = 10.85NDD150 pKa = 3.71DD151 pKa = 3.63GSRR154 pKa = 11.84NFGANVANSAVYY166 pKa = 10.21SVSQSFLVSCPSDD179 pKa = 3.15LGGTYY184 pKa = 10.33NYY186 pKa = 8.33STTDD190 pKa = 2.87AYY192 pKa = 10.42EE193 pKa = 4.1PGGASVAGPITGTVTFTAIGGGAYY217 pKa = 9.21EE218 pKa = 4.54ISDD221 pKa = 3.38ASFGGWEE228 pKa = 4.42AIYY231 pKa = 10.6GPGNISEE238 pKa = 4.57GVTLVDD244 pKa = 3.2VCNTIQFGGNDD255 pKa = 3.74QFGDD259 pKa = 3.9TYY261 pKa = 11.32TMSNLVVNGASLSFSWTNTYY281 pKa = 11.16GEE283 pKa = 4.55GGEE286 pKa = 4.4VTLTRR291 pKa = 11.84PDD293 pKa = 3.57GSPWPPLSLL302 pKa = 4.52
MM1 pKa = 7.58KK2 pKa = 10.36KK3 pKa = 9.42IQLFFLALSTALTLSGCEE21 pKa = 3.99TDD23 pKa = 5.45GGDD26 pKa = 4.15SKK28 pKa = 11.84LNLIEE33 pKa = 4.66AAVPDD38 pKa = 3.56ITINDD43 pKa = 3.76TSDD46 pKa = 3.43EE47 pKa = 4.52IINLVAVTNGGDD59 pKa = 3.31INLSLTLDD67 pKa = 3.31KK68 pKa = 11.55GFGKK72 pKa = 10.27VSSMDD77 pKa = 3.25VVLFYY82 pKa = 11.11YY83 pKa = 10.58KK84 pKa = 10.38SDD86 pKa = 3.06GSVFRR91 pKa = 11.84SVLQSGITEE100 pKa = 4.6FPATVSITKK109 pKa = 10.45NSIFDD114 pKa = 3.42AFEE117 pKa = 4.24QINVPADD124 pKa = 3.59FEE126 pKa = 4.71VGDD129 pKa = 4.34EE130 pKa = 4.23LLITADD136 pKa = 3.43LTLPNGTVIKK146 pKa = 9.75MYY148 pKa = 10.85NDD150 pKa = 3.71DD151 pKa = 3.63GSRR154 pKa = 11.84NFGANVANSAVYY166 pKa = 10.21SVSQSFLVSCPSDD179 pKa = 3.15LGGTYY184 pKa = 10.33NYY186 pKa = 8.33STTDD190 pKa = 2.87AYY192 pKa = 10.42EE193 pKa = 4.1PGGASVAGPITGTVTFTAIGGGAYY217 pKa = 9.21EE218 pKa = 4.54ISDD221 pKa = 3.38ASFGGWEE228 pKa = 4.42AIYY231 pKa = 10.6GPGNISEE238 pKa = 4.57GVTLVDD244 pKa = 3.2VCNTIQFGGNDD255 pKa = 3.74QFGDD259 pKa = 3.9TYY261 pKa = 11.32TMSNLVVNGASLSFSWTNTYY281 pKa = 11.16GEE283 pKa = 4.55GGEE286 pKa = 4.4VTLTRR291 pKa = 11.84PDD293 pKa = 3.57GSPWPPLSLL302 pKa = 4.52
Molecular weight: 31.67 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A255ZQJ0|A0A255ZQJ0_9FLAO Translation initiation factor SUI1-related protein OS=Flavobacterium aurantiibacter OX=2023067 GN=CHX27_08580 PE=3 SV=1
MM1 pKa = 7.65HH2 pKa = 7.53IFAQGVAAEE11 pKa = 4.43SRR13 pKa = 11.84AEE15 pKa = 4.05RR16 pKa = 11.84KK17 pKa = 8.8PVQPAAVRR25 pKa = 11.84RR26 pKa = 11.84PTEE29 pKa = 3.86AVPQFGGTVLEE40 pKa = 4.56RR41 pKa = 11.84CALQRR46 pKa = 11.84KK47 pKa = 5.44TRR49 pKa = 11.84PAALLYY55 pKa = 9.86FHH57 pKa = 7.45EE58 pKa = 5.08IFAAGCAQKK67 pKa = 9.74TGIDD71 pKa = 3.33TFTRR75 pKa = 11.84QITLLVCVPLLKK87 pKa = 10.25ILAHH91 pKa = 5.35VRR93 pKa = 11.84RR94 pKa = 4.37
MM1 pKa = 7.65HH2 pKa = 7.53IFAQGVAAEE11 pKa = 4.43SRR13 pKa = 11.84AEE15 pKa = 4.05RR16 pKa = 11.84KK17 pKa = 8.8PVQPAAVRR25 pKa = 11.84RR26 pKa = 11.84PTEE29 pKa = 3.86AVPQFGGTVLEE40 pKa = 4.56RR41 pKa = 11.84CALQRR46 pKa = 11.84KK47 pKa = 5.44TRR49 pKa = 11.84PAALLYY55 pKa = 9.86FHH57 pKa = 7.45EE58 pKa = 5.08IFAAGCAQKK67 pKa = 9.74TGIDD71 pKa = 3.33TFTRR75 pKa = 11.84QITLLVCVPLLKK87 pKa = 10.25ILAHH91 pKa = 5.35VRR93 pKa = 11.84RR94 pKa = 4.37
Molecular weight: 10.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
924509 |
34 |
3242 |
318.6 |
35.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.558 ± 0.056 | 0.851 ± 0.017 |
5.197 ± 0.033 | 6.162 ± 0.057 |
5.572 ± 0.039 | 6.245 ± 0.049 |
1.668 ± 0.023 | 7.201 ± 0.043 |
6.954 ± 0.076 | 9.468 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.921 ± 0.021 | 5.804 ± 0.05 |
3.577 ± 0.033 | 3.825 ± 0.026 |
3.933 ± 0.032 | 6.544 ± 0.044 |
6.092 ± 0.073 | 6.576 ± 0.041 |
0.979 ± 0.017 | 3.868 ± 0.032 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
