
Dishui Lake virophage 6
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Preplasmiviricota; Maveriviricetes; Priklausovirales; Lavidaviridae; unclassified Lavidaviridae
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
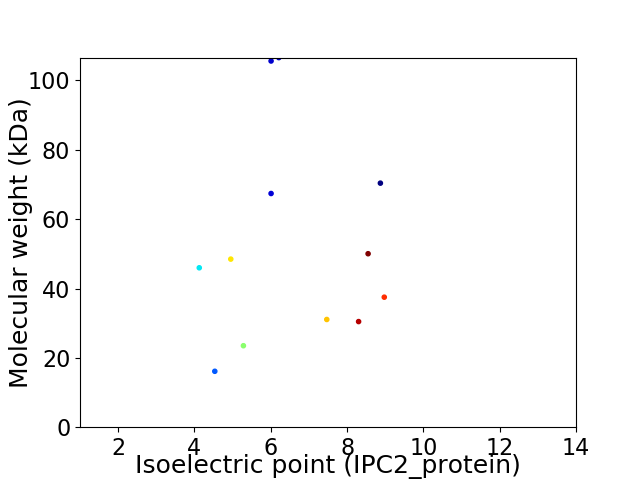
Virtual 2D-PAGE plot for 12 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6G6XMP1|A0A6G6XMP1_9VIRU Putative cysteine protease OS=Dishui Lake virophage 6 OX=2704067 PE=4 SV=1
MM1 pKa = 7.6SFDD4 pKa = 3.75TFCGQNRR11 pKa = 11.84TDD13 pKa = 3.27PSTYY17 pKa = 10.49QKK19 pKa = 11.23VLTKK23 pKa = 10.15FLSAAPTAVIDD34 pKa = 3.9TTNANATVFPVLAAGGGGLPQQLLVDD60 pKa = 5.07DD61 pKa = 4.96GLTPISVNCFNGNFNVVDD79 pKa = 3.92TLKK82 pKa = 9.25ITQTQVAVGKK92 pKa = 10.29GAGATNQAADD102 pKa = 3.78SVAVGIGAGTTNQGTQCVAVGEE124 pKa = 4.45ASGQSNQGANATAVGVVAGQINQGANATALGVNAGNSNQSANSVAVGLNAGEE176 pKa = 4.38TTQGADD182 pKa = 2.82SVAIGNVAGQTTQGASSVAIGINAGNTTQGASSVAVGSSAGQTTQGANSVAVGLNAGNTTQGVNATALGSNAANDD257 pKa = 3.8SQGNNGVAIGLNAGQTTQGADD278 pKa = 2.98AVAIGTNAGQTTQTLQAVAIGVNAGVTTQGANSVAIGAASGFTTQGANSVSIGLSSGTTNQGTQSVAVGADD349 pKa = 3.07SGGTNQGASSVAVGYY364 pKa = 8.88FAGGSAQGGSSVAIGVNAGRR384 pKa = 11.84TTQGDD389 pKa = 3.3NSVAVGGGAGQTTQGAGAVCIGSNAGNGNQGANAVAIGNLAGQTNQSAGSIVINASGAALNTGATVGLFIKK460 pKa = 9.74PIRR463 pKa = 11.84GVALGIGAGRR473 pKa = 11.84LFYY476 pKa = 10.88DD477 pKa = 3.71VATSEE482 pKa = 4.27IQYY485 pKa = 8.53STTT488 pKa = 3.12
MM1 pKa = 7.6SFDD4 pKa = 3.75TFCGQNRR11 pKa = 11.84TDD13 pKa = 3.27PSTYY17 pKa = 10.49QKK19 pKa = 11.23VLTKK23 pKa = 10.15FLSAAPTAVIDD34 pKa = 3.9TTNANATVFPVLAAGGGGLPQQLLVDD60 pKa = 5.07DD61 pKa = 4.96GLTPISVNCFNGNFNVVDD79 pKa = 3.92TLKK82 pKa = 9.25ITQTQVAVGKK92 pKa = 10.29GAGATNQAADD102 pKa = 3.78SVAVGIGAGTTNQGTQCVAVGEE124 pKa = 4.45ASGQSNQGANATAVGVVAGQINQGANATALGVNAGNSNQSANSVAVGLNAGEE176 pKa = 4.38TTQGADD182 pKa = 2.82SVAIGNVAGQTTQGASSVAIGINAGNTTQGASSVAVGSSAGQTTQGANSVAVGLNAGNTTQGVNATALGSNAANDD257 pKa = 3.8SQGNNGVAIGLNAGQTTQGADD278 pKa = 2.98AVAIGTNAGQTTQTLQAVAIGVNAGVTTQGANSVAIGAASGFTTQGANSVSIGLSSGTTNQGTQSVAVGADD349 pKa = 3.07SGGTNQGASSVAVGYY364 pKa = 8.88FAGGSAQGGSSVAIGVNAGRR384 pKa = 11.84TTQGDD389 pKa = 3.3NSVAVGGGAGQTTQGAGAVCIGSNAGNGNQGANAVAIGNLAGQTNQSAGSIVINASGAALNTGATVGLFIKK460 pKa = 9.74PIRR463 pKa = 11.84GVALGIGAGRR473 pKa = 11.84LFYY476 pKa = 10.88DD477 pKa = 3.71VATSEE482 pKa = 4.27IQYY485 pKa = 8.53STTT488 pKa = 3.12
Molecular weight: 45.98 kDa
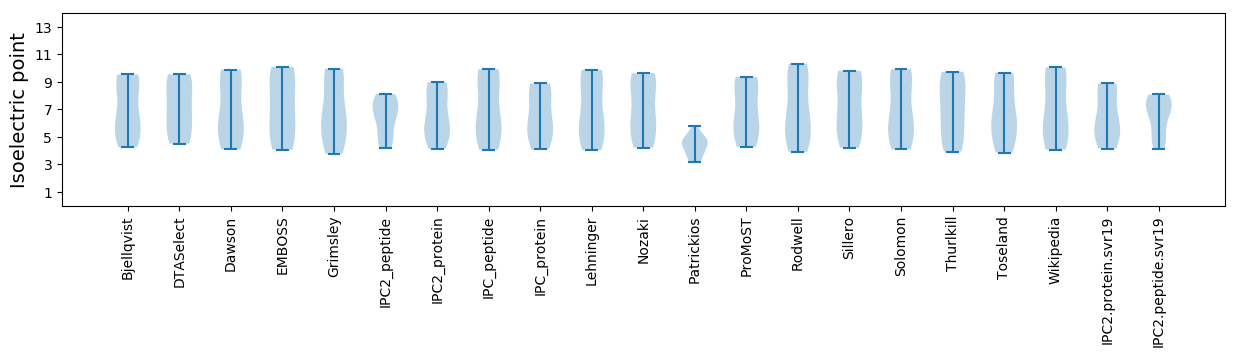
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6G6XMI1|A0A6G6XMI1_9VIRU Putative major capsid protein OS=Dishui Lake virophage 6 OX=2704067 PE=4 SV=1
MM1 pKa = 8.19DD2 pKa = 4.18FTSTLIEE9 pKa = 4.01KK10 pKa = 10.32LKK12 pKa = 10.8EE13 pKa = 3.94RR14 pKa = 11.84GLTDD18 pKa = 3.07SSVALYY24 pKa = 10.23VRR26 pKa = 11.84NLEE29 pKa = 4.34KK30 pKa = 11.04LNSNKK35 pKa = 9.68PLNNLNFLKK44 pKa = 10.29KK45 pKa = 10.88YY46 pKa = 9.87NDD48 pKa = 2.99IMEE51 pKa = 4.49KK52 pKa = 10.62LKK54 pKa = 10.78GYY56 pKa = 9.93KK57 pKa = 10.54GNTQRR62 pKa = 11.84GFLISIVSSLSSFKK76 pKa = 10.89GEE78 pKa = 4.13KK79 pKa = 10.68GIDD82 pKa = 3.6PLLKK86 pKa = 9.99KK87 pKa = 10.21YY88 pKa = 9.66YY89 pKa = 10.48KK90 pKa = 11.18AMIEE94 pKa = 4.2LNKK97 pKa = 10.52SLNEE101 pKa = 4.01ANHH104 pKa = 6.43NGLKK108 pKa = 9.71TEE110 pKa = 4.18TQSANWLDD118 pKa = 3.79WNDD121 pKa = 3.23VEE123 pKa = 5.86HH124 pKa = 7.59IYY126 pKa = 11.13DD127 pKa = 3.99GLRR130 pKa = 11.84DD131 pKa = 4.51NITQMSSPITEE142 pKa = 4.05GEE144 pKa = 4.13YY145 pKa = 10.89NRR147 pKa = 11.84LLDD150 pKa = 4.28LVVLSLYY157 pKa = 10.39VLNPPRR163 pKa = 11.84RR164 pKa = 11.84NSDD167 pKa = 3.56YY168 pKa = 10.78MNMKK172 pKa = 9.44VVSAFTPEE180 pKa = 3.82VSDD183 pKa = 4.07ALSGNNILDD192 pKa = 3.64WNGKK196 pKa = 8.69RR197 pKa = 11.84FIFRR201 pKa = 11.84NYY203 pKa = 8.17KK204 pKa = 6.84TAKK207 pKa = 10.0KK208 pKa = 9.37YY209 pKa = 11.2GEE211 pKa = 4.53TIVPIPRR218 pKa = 11.84EE219 pKa = 4.11LYY221 pKa = 10.57DD222 pKa = 3.27ILAVYY227 pKa = 10.05FDD229 pKa = 3.45KK230 pKa = 11.14RR231 pKa = 11.84GILRR235 pKa = 11.84RR236 pKa = 11.84LQVPQKK242 pKa = 8.85KK243 pKa = 7.31TKK245 pKa = 9.94KK246 pKa = 8.15EE247 pKa = 3.48ASIFIEE253 pKa = 4.44PFLTLWNDD261 pKa = 3.03KK262 pKa = 10.34PFLINTITRR271 pKa = 11.84ILNRR275 pKa = 11.84VFGKK279 pKa = 10.66KK280 pKa = 9.32IGSSMLRR287 pKa = 11.84HH288 pKa = 6.69IYY290 pKa = 6.2TTKK293 pKa = 10.92KK294 pKa = 10.15FGKK297 pKa = 9.61QLAEE301 pKa = 3.96QKK303 pKa = 8.52EE304 pKa = 4.34TAEE307 pKa = 4.23KK308 pKa = 9.32MGHH311 pKa = 5.79SVAEE315 pKa = 4.13MNQTYY320 pKa = 10.18IKK322 pKa = 10.52EE323 pKa = 4.17DD324 pKa = 3.15
MM1 pKa = 8.19DD2 pKa = 4.18FTSTLIEE9 pKa = 4.01KK10 pKa = 10.32LKK12 pKa = 10.8EE13 pKa = 3.94RR14 pKa = 11.84GLTDD18 pKa = 3.07SSVALYY24 pKa = 10.23VRR26 pKa = 11.84NLEE29 pKa = 4.34KK30 pKa = 11.04LNSNKK35 pKa = 9.68PLNNLNFLKK44 pKa = 10.29KK45 pKa = 10.88YY46 pKa = 9.87NDD48 pKa = 2.99IMEE51 pKa = 4.49KK52 pKa = 10.62LKK54 pKa = 10.78GYY56 pKa = 9.93KK57 pKa = 10.54GNTQRR62 pKa = 11.84GFLISIVSSLSSFKK76 pKa = 10.89GEE78 pKa = 4.13KK79 pKa = 10.68GIDD82 pKa = 3.6PLLKK86 pKa = 9.99KK87 pKa = 10.21YY88 pKa = 9.66YY89 pKa = 10.48KK90 pKa = 11.18AMIEE94 pKa = 4.2LNKK97 pKa = 10.52SLNEE101 pKa = 4.01ANHH104 pKa = 6.43NGLKK108 pKa = 9.71TEE110 pKa = 4.18TQSANWLDD118 pKa = 3.79WNDD121 pKa = 3.23VEE123 pKa = 5.86HH124 pKa = 7.59IYY126 pKa = 11.13DD127 pKa = 3.99GLRR130 pKa = 11.84DD131 pKa = 4.51NITQMSSPITEE142 pKa = 4.05GEE144 pKa = 4.13YY145 pKa = 10.89NRR147 pKa = 11.84LLDD150 pKa = 4.28LVVLSLYY157 pKa = 10.39VLNPPRR163 pKa = 11.84RR164 pKa = 11.84NSDD167 pKa = 3.56YY168 pKa = 10.78MNMKK172 pKa = 9.44VVSAFTPEE180 pKa = 3.82VSDD183 pKa = 4.07ALSGNNILDD192 pKa = 3.64WNGKK196 pKa = 8.69RR197 pKa = 11.84FIFRR201 pKa = 11.84NYY203 pKa = 8.17KK204 pKa = 6.84TAKK207 pKa = 10.0KK208 pKa = 9.37YY209 pKa = 11.2GEE211 pKa = 4.53TIVPIPRR218 pKa = 11.84EE219 pKa = 4.11LYY221 pKa = 10.57DD222 pKa = 3.27ILAVYY227 pKa = 10.05FDD229 pKa = 3.45KK230 pKa = 11.14RR231 pKa = 11.84GILRR235 pKa = 11.84RR236 pKa = 11.84LQVPQKK242 pKa = 8.85KK243 pKa = 7.31TKK245 pKa = 9.94KK246 pKa = 8.15EE247 pKa = 3.48ASIFIEE253 pKa = 4.44PFLTLWNDD261 pKa = 3.03KK262 pKa = 10.34PFLINTITRR271 pKa = 11.84ILNRR275 pKa = 11.84VFGKK279 pKa = 10.66KK280 pKa = 9.32IGSSMLRR287 pKa = 11.84HH288 pKa = 6.69IYY290 pKa = 6.2TTKK293 pKa = 10.92KK294 pKa = 10.15FGKK297 pKa = 9.61QLAEE301 pKa = 3.96QKK303 pKa = 8.52EE304 pKa = 4.34TAEE307 pKa = 4.23KK308 pKa = 9.32MGHH311 pKa = 5.79SVAEE315 pKa = 4.13MNQTYY320 pKa = 10.18IKK322 pKa = 10.52EE323 pKa = 4.17DD324 pKa = 3.15
Molecular weight: 37.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5679 |
143 |
952 |
473.3 |
52.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.924 ± 0.916 | 0.845 ± 0.199 |
5.494 ± 0.347 | 6.515 ± 0.825 |
3.751 ± 0.301 | 7.096 ± 0.958 |
1.109 ± 0.207 | 6.41 ± 0.543 |
6.057 ± 0.951 | 8.382 ± 0.42 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.06 ± 0.225 | 6.498 ± 0.543 |
5.089 ± 0.683 | 4.825 ± 0.87 |
5.917 ± 0.898 | 6.374 ± 0.675 |
5.23 ± 0.637 | 6.11 ± 0.415 |
0.898 ± 0.279 | 3.416 ± 0.337 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
