
Oita virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Ledantevirus; Oita ledantevirus
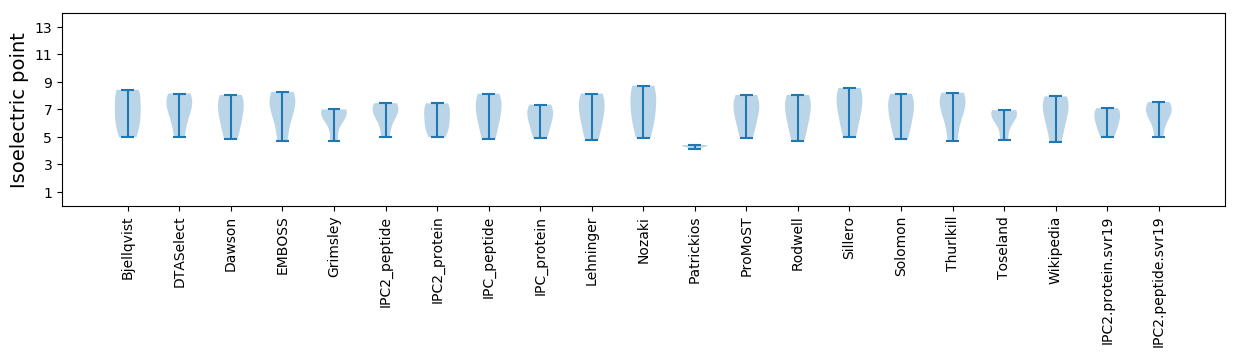
Average proteome isoelectric point is 6.21
Get precalculated fractions of proteins
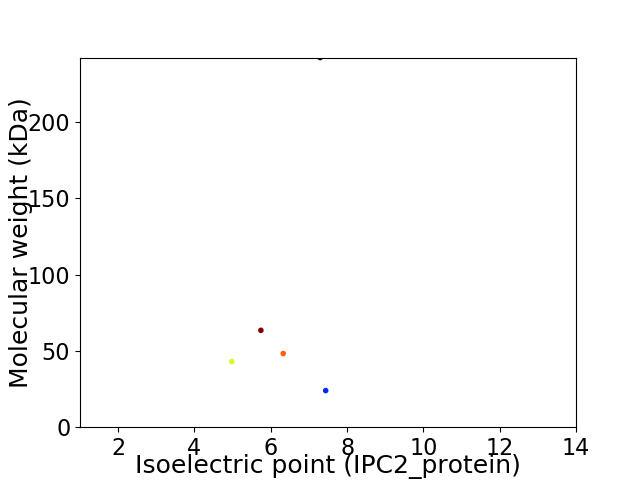
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0D3R163|A0A0D3R163_9RHAB Nucleocapsid protein OS=Oita virus OX=1272953 PE=4 SV=1
MM1 pKa = 7.35NSDD4 pKa = 3.24RR5 pKa = 11.84AKK7 pKa = 10.03IQRR10 pKa = 11.84LAKK13 pKa = 9.91ALPWEE18 pKa = 4.04NVKK21 pKa = 10.6INIEE25 pKa = 4.38DD26 pKa = 3.76IEE28 pKa = 4.59DD29 pKa = 3.71SEE31 pKa = 5.02EE32 pKa = 4.39EE33 pKa = 3.81IDD35 pKa = 3.54QKK37 pKa = 11.16IIVTEE42 pKa = 3.98EE43 pKa = 3.98RR44 pKa = 11.84DD45 pKa = 4.17LEE47 pKa = 4.34WTEE50 pKa = 4.16NPIKK54 pKa = 10.53EE55 pKa = 4.13QVEE58 pKa = 3.86PFEE61 pKa = 4.22IQEE64 pKa = 4.16EE65 pKa = 4.52DD66 pKa = 3.53STPTEE71 pKa = 4.31MGLSACEE78 pKa = 3.9KK79 pKa = 9.56EE80 pKa = 4.22KK81 pKa = 11.09EE82 pKa = 4.11KK83 pKa = 11.16EE84 pKa = 4.14KK85 pKa = 11.28EE86 pKa = 3.58KK87 pKa = 10.67DD88 pKa = 3.01TAEE91 pKa = 3.99NKK93 pKa = 10.46GIEE96 pKa = 4.37GSGQGCSKK104 pKa = 10.13RR105 pKa = 11.84SKK107 pKa = 9.98RR108 pKa = 11.84GKK110 pKa = 10.05GEE112 pKa = 3.6QRR114 pKa = 11.84GKK116 pKa = 10.47NKK118 pKa = 10.26SEE120 pKa = 4.0EE121 pKa = 3.99TSAPSDD127 pKa = 3.28SATITNPDD135 pKa = 3.07IKK137 pKa = 10.39EE138 pKa = 4.32DD139 pKa = 3.95PVCALSVEE147 pKa = 4.52EE148 pKa = 3.98DD149 pKa = 3.58ALWRR153 pKa = 11.84EE154 pKa = 4.59EE155 pKa = 3.82KK156 pKa = 10.47RR157 pKa = 11.84KK158 pKa = 9.46EE159 pKa = 3.96ALRR162 pKa = 11.84LQSDD166 pKa = 4.87AIRR169 pKa = 11.84VEE171 pKa = 3.77RR172 pKa = 11.84VNYY175 pKa = 10.57NEE177 pKa = 4.98IKK179 pKa = 9.01TQEE182 pKa = 4.31DD183 pKa = 4.02LDD185 pKa = 4.24DD186 pKa = 5.45LMTEE190 pKa = 4.36VIYY193 pKa = 10.72KK194 pKa = 10.16VLEE197 pKa = 4.12CTGYY201 pKa = 10.44CARR204 pKa = 11.84SDD206 pKa = 3.53LTSIRR211 pKa = 11.84RR212 pKa = 11.84DD213 pKa = 3.92DD214 pKa = 3.1IRR216 pKa = 11.84VYY218 pKa = 10.76YY219 pKa = 10.37KK220 pKa = 10.62EE221 pKa = 4.55SFVKK225 pKa = 9.91EE226 pKa = 4.07AEE228 pKa = 4.24RR229 pKa = 11.84QTEE232 pKa = 4.06EE233 pKa = 3.4WKK235 pKa = 10.68KK236 pKa = 10.64RR237 pKa = 11.84LEE239 pKa = 4.0EE240 pKa = 4.29EE241 pKa = 4.95IKK243 pKa = 11.07DD244 pKa = 3.82KK245 pKa = 10.59TCKK248 pKa = 10.49DD249 pKa = 3.75NLCDD253 pKa = 4.01PDD255 pKa = 4.14SKK257 pKa = 11.1RR258 pKa = 11.84EE259 pKa = 3.92EE260 pKa = 3.98NKK262 pKa = 10.69ANPKK266 pKa = 9.63DD267 pKa = 3.82QSSSSIKK274 pKa = 9.79EE275 pKa = 3.53RR276 pKa = 11.84HH277 pKa = 4.8QPQIEE282 pKa = 4.54STDD285 pKa = 3.39EE286 pKa = 3.92PNPRR290 pKa = 11.84KK291 pKa = 9.49EE292 pKa = 4.08DD293 pKa = 3.45RR294 pKa = 11.84SDD296 pKa = 3.37NAEE299 pKa = 4.44DD300 pKa = 3.34STAIQMQLEE309 pKa = 4.3TLLQEE314 pKa = 4.54GMCFRR319 pKa = 11.84KK320 pKa = 10.0KK321 pKa = 10.07RR322 pKa = 11.84GGYY325 pKa = 6.59MTVSHH330 pKa = 7.62DD331 pKa = 3.64NPKK334 pKa = 10.61LKK336 pKa = 10.49NIKK339 pKa = 9.93LKK341 pKa = 10.71EE342 pKa = 3.89IALSSGSVKK351 pKa = 9.62EE352 pKa = 4.33AKK354 pKa = 9.91ILLLKK359 pKa = 10.6KK360 pKa = 10.26AGLYY364 pKa = 10.0QGIKK368 pKa = 9.3LLCYY372 pKa = 9.89IDD374 pKa = 3.71
MM1 pKa = 7.35NSDD4 pKa = 3.24RR5 pKa = 11.84AKK7 pKa = 10.03IQRR10 pKa = 11.84LAKK13 pKa = 9.91ALPWEE18 pKa = 4.04NVKK21 pKa = 10.6INIEE25 pKa = 4.38DD26 pKa = 3.76IEE28 pKa = 4.59DD29 pKa = 3.71SEE31 pKa = 5.02EE32 pKa = 4.39EE33 pKa = 3.81IDD35 pKa = 3.54QKK37 pKa = 11.16IIVTEE42 pKa = 3.98EE43 pKa = 3.98RR44 pKa = 11.84DD45 pKa = 4.17LEE47 pKa = 4.34WTEE50 pKa = 4.16NPIKK54 pKa = 10.53EE55 pKa = 4.13QVEE58 pKa = 3.86PFEE61 pKa = 4.22IQEE64 pKa = 4.16EE65 pKa = 4.52DD66 pKa = 3.53STPTEE71 pKa = 4.31MGLSACEE78 pKa = 3.9KK79 pKa = 9.56EE80 pKa = 4.22KK81 pKa = 11.09EE82 pKa = 4.11KK83 pKa = 11.16EE84 pKa = 4.14KK85 pKa = 11.28EE86 pKa = 3.58KK87 pKa = 10.67DD88 pKa = 3.01TAEE91 pKa = 3.99NKK93 pKa = 10.46GIEE96 pKa = 4.37GSGQGCSKK104 pKa = 10.13RR105 pKa = 11.84SKK107 pKa = 9.98RR108 pKa = 11.84GKK110 pKa = 10.05GEE112 pKa = 3.6QRR114 pKa = 11.84GKK116 pKa = 10.47NKK118 pKa = 10.26SEE120 pKa = 4.0EE121 pKa = 3.99TSAPSDD127 pKa = 3.28SATITNPDD135 pKa = 3.07IKK137 pKa = 10.39EE138 pKa = 4.32DD139 pKa = 3.95PVCALSVEE147 pKa = 4.52EE148 pKa = 3.98DD149 pKa = 3.58ALWRR153 pKa = 11.84EE154 pKa = 4.59EE155 pKa = 3.82KK156 pKa = 10.47RR157 pKa = 11.84KK158 pKa = 9.46EE159 pKa = 3.96ALRR162 pKa = 11.84LQSDD166 pKa = 4.87AIRR169 pKa = 11.84VEE171 pKa = 3.77RR172 pKa = 11.84VNYY175 pKa = 10.57NEE177 pKa = 4.98IKK179 pKa = 9.01TQEE182 pKa = 4.31DD183 pKa = 4.02LDD185 pKa = 4.24DD186 pKa = 5.45LMTEE190 pKa = 4.36VIYY193 pKa = 10.72KK194 pKa = 10.16VLEE197 pKa = 4.12CTGYY201 pKa = 10.44CARR204 pKa = 11.84SDD206 pKa = 3.53LTSIRR211 pKa = 11.84RR212 pKa = 11.84DD213 pKa = 3.92DD214 pKa = 3.1IRR216 pKa = 11.84VYY218 pKa = 10.76YY219 pKa = 10.37KK220 pKa = 10.62EE221 pKa = 4.55SFVKK225 pKa = 9.91EE226 pKa = 4.07AEE228 pKa = 4.24RR229 pKa = 11.84QTEE232 pKa = 4.06EE233 pKa = 3.4WKK235 pKa = 10.68KK236 pKa = 10.64RR237 pKa = 11.84LEE239 pKa = 4.0EE240 pKa = 4.29EE241 pKa = 4.95IKK243 pKa = 11.07DD244 pKa = 3.82KK245 pKa = 10.59TCKK248 pKa = 10.49DD249 pKa = 3.75NLCDD253 pKa = 4.01PDD255 pKa = 4.14SKK257 pKa = 11.1RR258 pKa = 11.84EE259 pKa = 3.92EE260 pKa = 3.98NKK262 pKa = 10.69ANPKK266 pKa = 9.63DD267 pKa = 3.82QSSSSIKK274 pKa = 9.79EE275 pKa = 3.53RR276 pKa = 11.84HH277 pKa = 4.8QPQIEE282 pKa = 4.54STDD285 pKa = 3.39EE286 pKa = 3.92PNPRR290 pKa = 11.84KK291 pKa = 9.49EE292 pKa = 4.08DD293 pKa = 3.45RR294 pKa = 11.84SDD296 pKa = 3.37NAEE299 pKa = 4.44DD300 pKa = 3.34STAIQMQLEE309 pKa = 4.3TLLQEE314 pKa = 4.54GMCFRR319 pKa = 11.84KK320 pKa = 10.0KK321 pKa = 10.07RR322 pKa = 11.84GGYY325 pKa = 6.59MTVSHH330 pKa = 7.62DD331 pKa = 3.64NPKK334 pKa = 10.61LKK336 pKa = 10.49NIKK339 pKa = 9.93LKK341 pKa = 10.71EE342 pKa = 3.89IALSSGSVKK351 pKa = 9.62EE352 pKa = 4.33AKK354 pKa = 9.91ILLLKK359 pKa = 10.6KK360 pKa = 10.26AGLYY364 pKa = 10.0QGIKK368 pKa = 9.3LLCYY372 pKa = 9.89IDD374 pKa = 3.71
Molecular weight: 43.08 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0D3R1Q4|A0A0D3R1Q4_9RHAB Matrix OS=Oita virus OX=1272953 PE=4 SV=1
MM1 pKa = 7.47LSVFKK6 pKa = 10.59KK7 pKa = 9.77SKK9 pKa = 9.68KK10 pKa = 10.05DD11 pKa = 3.5RR12 pKa = 11.84VSSSAPDD19 pKa = 3.32TSQLVKK25 pKa = 9.71EE26 pKa = 4.74TFFSVPSAPAPSIKK40 pKa = 9.96KK41 pKa = 10.42DD42 pKa = 3.16IFTHH46 pKa = 4.89TLRR49 pKa = 11.84VQAWIEE55 pKa = 3.8IRR57 pKa = 11.84SEE59 pKa = 3.77EE60 pKa = 4.29SIQDD64 pKa = 3.65FEE66 pKa = 5.19DD67 pKa = 3.48CTKK70 pKa = 10.45ILEE73 pKa = 4.14PWIDD77 pKa = 3.74EE78 pKa = 4.17ASCPIWQYY86 pKa = 11.3IIDD89 pKa = 3.8SWVFFCMGVHH99 pKa = 6.26ARR101 pKa = 11.84KK102 pKa = 10.13DD103 pKa = 3.77PLCTTHH109 pKa = 6.75CLYY112 pKa = 10.51RR113 pKa = 11.84ARR115 pKa = 11.84IDD117 pKa = 3.3QVLEE121 pKa = 4.33FKK123 pKa = 11.21HH124 pKa = 6.2NVEE127 pKa = 4.35EE128 pKa = 5.44LDD130 pKa = 3.69PALCKK135 pKa = 10.38AVTVRR140 pKa = 11.84FVTAYY145 pKa = 10.17NGKK148 pKa = 9.02KK149 pKa = 10.14CEE151 pKa = 3.88VSFWSSMTPSRR162 pKa = 11.84RR163 pKa = 11.84RR164 pKa = 11.84GAPFFIMYY172 pKa = 7.5YY173 pKa = 10.87APLKK177 pKa = 10.76DD178 pKa = 4.3GNPPPAMMKK187 pKa = 9.99WKK189 pKa = 10.27INLPFKK195 pKa = 10.5ILNDD199 pKa = 4.0TEE201 pKa = 4.06EE202 pKa = 4.6GPYY205 pKa = 10.27LSKK208 pKa = 11.23
MM1 pKa = 7.47LSVFKK6 pKa = 10.59KK7 pKa = 9.77SKK9 pKa = 9.68KK10 pKa = 10.05DD11 pKa = 3.5RR12 pKa = 11.84VSSSAPDD19 pKa = 3.32TSQLVKK25 pKa = 9.71EE26 pKa = 4.74TFFSVPSAPAPSIKK40 pKa = 9.96KK41 pKa = 10.42DD42 pKa = 3.16IFTHH46 pKa = 4.89TLRR49 pKa = 11.84VQAWIEE55 pKa = 3.8IRR57 pKa = 11.84SEE59 pKa = 3.77EE60 pKa = 4.29SIQDD64 pKa = 3.65FEE66 pKa = 5.19DD67 pKa = 3.48CTKK70 pKa = 10.45ILEE73 pKa = 4.14PWIDD77 pKa = 3.74EE78 pKa = 4.17ASCPIWQYY86 pKa = 11.3IIDD89 pKa = 3.8SWVFFCMGVHH99 pKa = 6.26ARR101 pKa = 11.84KK102 pKa = 10.13DD103 pKa = 3.77PLCTTHH109 pKa = 6.75CLYY112 pKa = 10.51RR113 pKa = 11.84ARR115 pKa = 11.84IDD117 pKa = 3.3QVLEE121 pKa = 4.33FKK123 pKa = 11.21HH124 pKa = 6.2NVEE127 pKa = 4.35EE128 pKa = 5.44LDD130 pKa = 3.69PALCKK135 pKa = 10.38AVTVRR140 pKa = 11.84FVTAYY145 pKa = 10.17NGKK148 pKa = 9.02KK149 pKa = 10.14CEE151 pKa = 3.88VSFWSSMTPSRR162 pKa = 11.84RR163 pKa = 11.84RR164 pKa = 11.84GAPFFIMYY172 pKa = 7.5YY173 pKa = 10.87APLKK177 pKa = 10.76DD178 pKa = 4.3GNPPPAMMKK187 pKa = 9.99WKK189 pKa = 10.27INLPFKK195 pKa = 10.5ILNDD199 pKa = 4.0TEE201 pKa = 4.06EE202 pKa = 4.6GPYY205 pKa = 10.27LSKK208 pKa = 11.23
Molecular weight: 24.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3682 |
208 |
2119 |
736.4 |
84.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.726 ± 0.83 | 2.146 ± 0.23 |
5.703 ± 0.397 | 7.74 ± 1.503 |
4.047 ± 0.61 | 6.165 ± 0.796 |
2.499 ± 0.47 | 7.17 ± 0.596 |
7.251 ± 1.04 | 9.533 ± 1.103 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.146 ± 0.152 | 4.59 ± 0.454 |
4.237 ± 0.419 | 3.069 ± 0.331 |
5.731 ± 0.398 | 7.414 ± 0.291 |
4.916 ± 0.395 | 5.595 ± 0.537 |
2.01 ± 0.182 | 3.313 ± 0.262 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
