
Delftia phage RG-2014
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Schitoviridae; Dendoorenvirus; Delftia virus RG2014
Average proteome isoelectric point is 6.58
Get precalculated fractions of proteins
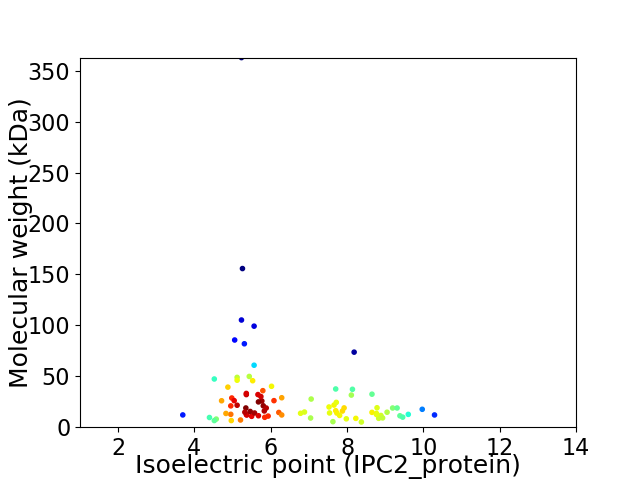
Virtual 2D-PAGE plot for 88 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
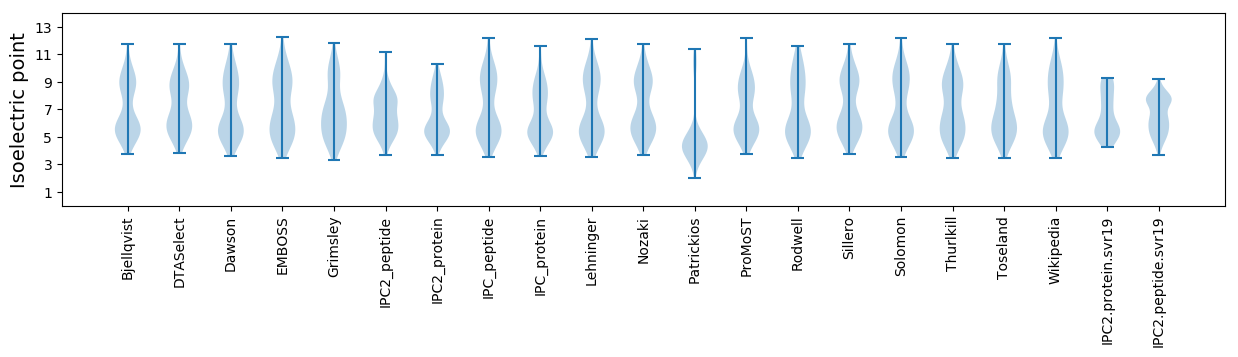
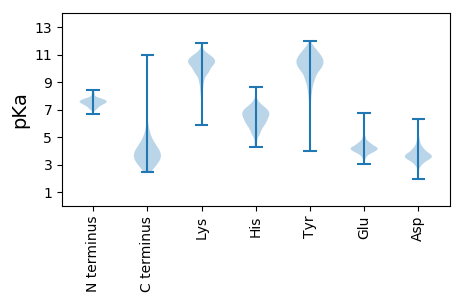
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A097PBE4|A0A097PBE4_9CAUD Uncharacterized protein OS=Delftia phage RG-2014 OX=1563661 GN=RG2014_087 PE=4 SV=1
MM1 pKa = 7.29SVISVIPEE9 pKa = 3.76PDD11 pKa = 3.32FNAQKK16 pKa = 10.94AGIRR20 pKa = 11.84QLQAEE25 pKa = 4.98LAAEE29 pKa = 4.08MSDD32 pKa = 3.91VIVGAQTTQYY42 pKa = 11.22GPDD45 pKa = 3.49EE46 pKa = 4.36YY47 pKa = 10.82TFEE50 pKa = 5.24PGLGQIHH57 pKa = 5.42YY58 pKa = 7.68TAATANKK65 pKa = 9.3IGKK68 pKa = 9.19DD69 pKa = 3.44LGASLAAALGNVYY82 pKa = 10.19PNPPTLKK89 pKa = 10.29PNPDD93 pKa = 4.36DD94 pKa = 3.38YY95 pKa = 11.48WFMGEE100 pKa = 5.29LYY102 pKa = 10.22PEE104 pKa = 4.86ADD106 pKa = 3.74FEE108 pKa = 4.62TEE110 pKa = 4.07PVNVATMDD118 pKa = 3.43YY119 pKa = 10.44YY120 pKa = 11.25RR121 pKa = 11.84RR122 pKa = 11.84FLKK125 pKa = 10.51LLHH128 pKa = 5.86EE129 pKa = 4.64RR130 pKa = 11.84EE131 pKa = 4.35FKK133 pKa = 10.6FVNSVAYY140 pKa = 9.7EE141 pKa = 3.84ILDD144 pKa = 3.85FFHH147 pKa = 7.32PEE149 pKa = 3.04AWKK152 pKa = 10.21QRR154 pKa = 11.84NFLGNPGLSGWYY166 pKa = 8.69PPSAFIQPTNFDD178 pKa = 3.55ALNYY182 pKa = 9.51ISAVQAQILRR192 pKa = 11.84EE193 pKa = 3.98AVALGMEE200 pKa = 4.56PLFQIGEE207 pKa = 4.2PWWWDD212 pKa = 3.02GSYY215 pKa = 9.24STGEE219 pKa = 4.22GKK221 pKa = 9.6NAPCLYY227 pKa = 10.64DD228 pKa = 3.61PATMVLYY235 pKa = 10.11EE236 pKa = 4.81AEE238 pKa = 4.38TGNQVPTPYY247 pKa = 9.63IKK249 pKa = 10.76SIFDD253 pKa = 3.75PVASNQWPYY262 pKa = 11.39VDD264 pKa = 3.79WLRR267 pKa = 11.84KK268 pKa = 9.67KK269 pKa = 10.91LGDD272 pKa = 3.4STNYY276 pKa = 9.35IRR278 pKa = 11.84DD279 pKa = 3.65RR280 pKa = 11.84VKK282 pKa = 11.09SLIPGSKK289 pKa = 8.97ATLLFFTPQIMSPASEE305 pKa = 3.76LTYY308 pKa = 10.89RR309 pKa = 11.84LNFPIAEE316 pKa = 3.94WSYY319 pKa = 9.95PNYY322 pKa = 10.79DD323 pKa = 3.16FVQIEE328 pKa = 4.35DD329 pKa = 4.13YY330 pKa = 11.26DD331 pKa = 3.96WIIAGDD337 pKa = 3.6IEE339 pKa = 5.1LVPKK343 pKa = 10.35TFDD346 pKa = 3.65AATNLLNYY354 pKa = 9.01PLSEE358 pKa = 3.63VHH360 pKa = 5.8YY361 pKa = 10.18FIGFVLKK368 pKa = 11.07AEE370 pKa = 4.57DD371 pKa = 3.57AYY373 pKa = 10.02IWRR376 pKa = 11.84TCDD379 pKa = 3.05QALGLAIEE387 pKa = 4.08AQIPYY392 pKa = 9.83KK393 pKa = 10.3YY394 pKa = 9.62LWSYY398 pKa = 6.96TQVMRR403 pKa = 11.84DD404 pKa = 3.74NIVMQRR410 pKa = 11.84LRR412 pKa = 11.84DD413 pKa = 3.88CGCC416 pKa = 3.68
MM1 pKa = 7.29SVISVIPEE9 pKa = 3.76PDD11 pKa = 3.32FNAQKK16 pKa = 10.94AGIRR20 pKa = 11.84QLQAEE25 pKa = 4.98LAAEE29 pKa = 4.08MSDD32 pKa = 3.91VIVGAQTTQYY42 pKa = 11.22GPDD45 pKa = 3.49EE46 pKa = 4.36YY47 pKa = 10.82TFEE50 pKa = 5.24PGLGQIHH57 pKa = 5.42YY58 pKa = 7.68TAATANKK65 pKa = 9.3IGKK68 pKa = 9.19DD69 pKa = 3.44LGASLAAALGNVYY82 pKa = 10.19PNPPTLKK89 pKa = 10.29PNPDD93 pKa = 4.36DD94 pKa = 3.38YY95 pKa = 11.48WFMGEE100 pKa = 5.29LYY102 pKa = 10.22PEE104 pKa = 4.86ADD106 pKa = 3.74FEE108 pKa = 4.62TEE110 pKa = 4.07PVNVATMDD118 pKa = 3.43YY119 pKa = 10.44YY120 pKa = 11.25RR121 pKa = 11.84RR122 pKa = 11.84FLKK125 pKa = 10.51LLHH128 pKa = 5.86EE129 pKa = 4.64RR130 pKa = 11.84EE131 pKa = 4.35FKK133 pKa = 10.6FVNSVAYY140 pKa = 9.7EE141 pKa = 3.84ILDD144 pKa = 3.85FFHH147 pKa = 7.32PEE149 pKa = 3.04AWKK152 pKa = 10.21QRR154 pKa = 11.84NFLGNPGLSGWYY166 pKa = 8.69PPSAFIQPTNFDD178 pKa = 3.55ALNYY182 pKa = 9.51ISAVQAQILRR192 pKa = 11.84EE193 pKa = 3.98AVALGMEE200 pKa = 4.56PLFQIGEE207 pKa = 4.2PWWWDD212 pKa = 3.02GSYY215 pKa = 9.24STGEE219 pKa = 4.22GKK221 pKa = 9.6NAPCLYY227 pKa = 10.64DD228 pKa = 3.61PATMVLYY235 pKa = 10.11EE236 pKa = 4.81AEE238 pKa = 4.38TGNQVPTPYY247 pKa = 9.63IKK249 pKa = 10.76SIFDD253 pKa = 3.75PVASNQWPYY262 pKa = 11.39VDD264 pKa = 3.79WLRR267 pKa = 11.84KK268 pKa = 9.67KK269 pKa = 10.91LGDD272 pKa = 3.4STNYY276 pKa = 9.35IRR278 pKa = 11.84DD279 pKa = 3.65RR280 pKa = 11.84VKK282 pKa = 11.09SLIPGSKK289 pKa = 8.97ATLLFFTPQIMSPASEE305 pKa = 3.76LTYY308 pKa = 10.89RR309 pKa = 11.84LNFPIAEE316 pKa = 3.94WSYY319 pKa = 9.95PNYY322 pKa = 10.79DD323 pKa = 3.16FVQIEE328 pKa = 4.35DD329 pKa = 4.13YY330 pKa = 11.26DD331 pKa = 3.96WIIAGDD337 pKa = 3.6IEE339 pKa = 5.1LVPKK343 pKa = 10.35TFDD346 pKa = 3.65AATNLLNYY354 pKa = 9.01PLSEE358 pKa = 3.63VHH360 pKa = 5.8YY361 pKa = 10.18FIGFVLKK368 pKa = 11.07AEE370 pKa = 4.57DD371 pKa = 3.57AYY373 pKa = 10.02IWRR376 pKa = 11.84TCDD379 pKa = 3.05QALGLAIEE387 pKa = 4.08AQIPYY392 pKa = 9.83KK393 pKa = 10.3YY394 pKa = 9.62LWSYY398 pKa = 6.96TQVMRR403 pKa = 11.84DD404 pKa = 3.74NIVMQRR410 pKa = 11.84LRR412 pKa = 11.84DD413 pKa = 3.88CGCC416 pKa = 3.68
Molecular weight: 47.32 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A097PAK7|A0A097PAK7_9CAUD Uncharacterized protein OS=Delftia phage RG-2014 OX=1563661 GN=RG2014_011 PE=4 SV=1
MM1 pKa = 7.17AQFYY5 pKa = 8.27VQPLHH10 pKa = 6.38NPNSGEE16 pKa = 3.83FCTVLRR22 pKa = 11.84QMRR25 pKa = 11.84HH26 pKa = 5.06LSMLGITVPKK36 pKa = 8.18TLKK39 pKa = 10.26VLATGRR45 pKa = 11.84VEE47 pKa = 5.24GYY49 pKa = 10.52QYY51 pKa = 10.08TCPSSRR57 pKa = 11.84RR58 pKa = 11.84QAQINEE64 pKa = 4.59TPVHH68 pKa = 5.98MGRR71 pKa = 11.84FSIEE75 pKa = 3.51RR76 pKa = 11.84VQTRR80 pKa = 11.84KK81 pKa = 8.84GTWLRR86 pKa = 11.84YY87 pKa = 10.24VNIAWGGEE95 pKa = 3.59IHH97 pKa = 7.1RR98 pKa = 11.84VQIQPTTLFIDD109 pKa = 3.93
MM1 pKa = 7.17AQFYY5 pKa = 8.27VQPLHH10 pKa = 6.38NPNSGEE16 pKa = 3.83FCTVLRR22 pKa = 11.84QMRR25 pKa = 11.84HH26 pKa = 5.06LSMLGITVPKK36 pKa = 8.18TLKK39 pKa = 10.26VLATGRR45 pKa = 11.84VEE47 pKa = 5.24GYY49 pKa = 10.52QYY51 pKa = 10.08TCPSSRR57 pKa = 11.84RR58 pKa = 11.84QAQINEE64 pKa = 4.59TPVHH68 pKa = 5.98MGRR71 pKa = 11.84FSIEE75 pKa = 3.51RR76 pKa = 11.84VQTRR80 pKa = 11.84KK81 pKa = 8.84GTWLRR86 pKa = 11.84YY87 pKa = 10.24VNIAWGGEE95 pKa = 3.59IHH97 pKa = 7.1RR98 pKa = 11.84VQIQPTTLFIDD109 pKa = 3.93
Molecular weight: 12.59 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
23589 |
44 |
3413 |
268.1 |
29.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.17 ± 0.745 | 0.945 ± 0.135 |
5.918 ± 0.13 | 6.066 ± 0.17 |
3.599 ± 0.174 | 7.092 ± 0.245 |
2.022 ± 0.174 | 4.812 ± 0.226 |
5.494 ± 0.153 | 8.474 ± 0.229 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.345 ± 0.107 | 4.239 ± 0.174 |
4.752 ± 0.205 | 4.943 ± 0.236 |
5.32 ± 0.199 | 5.193 ± 0.142 |
5.558 ± 0.197 | 6.291 ± 0.247 |
1.662 ± 0.178 | 3.103 ± 0.224 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
