
Nocardioides sp. CF204
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Propionibacteriales; Nocardioidaceae; Nocardioides; unclassified Nocardioides
Average proteome isoelectric point is 6.03
Get precalculated fractions of proteins
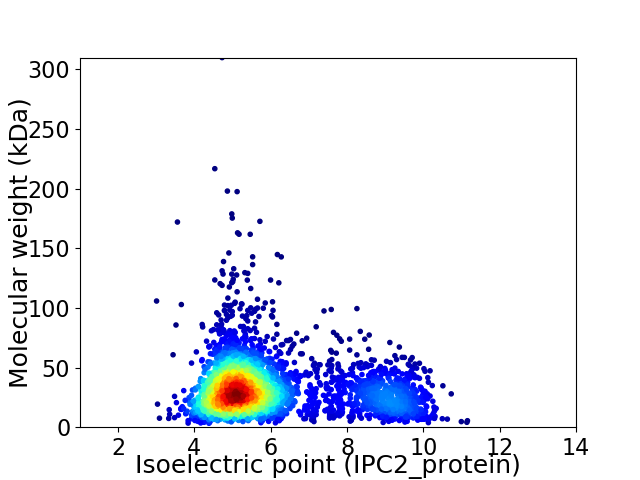
Virtual 2D-PAGE plot for 3302 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2T5W9M8|A0A2T5W9M8_9ACTN Single-strand DNA-binding protein OS=Nocardioides sp. CF204 OX=2135677 GN=C8J51_2070 PE=4 SV=1
MM1 pKa = 7.97RR2 pKa = 11.84PPFLRR7 pKa = 11.84TTLAGMLVAATLGLAPAAFADD28 pKa = 3.71QTSRR32 pKa = 11.84DD33 pKa = 3.45GDD35 pKa = 3.76TAAVGRR41 pKa = 11.84NITYY45 pKa = 10.22GPGFRR50 pKa = 11.84ACTTRR55 pKa = 11.84GAQVSGAITVTYY67 pKa = 10.78NGGTHH72 pKa = 6.17LTPGEE77 pKa = 3.98ALVVTMSPPAGVTATAGATSVPATWNSNGQSFVIPLSTTVTTAAASGSVGFKK129 pKa = 10.68VVGASSGYY137 pKa = 10.57DD138 pKa = 3.22VTAGGSQYY146 pKa = 11.47DD147 pKa = 4.08VIVDD151 pKa = 4.28CPANTAPTGVADD163 pKa = 5.17SYY165 pKa = 11.67ATDD168 pKa = 3.42EE169 pKa = 4.32DD170 pKa = 4.29TAKK173 pKa = 10.6VVAAPGVLGNDD184 pKa = 3.6TDD186 pKa = 4.09PQGDD190 pKa = 3.94ALTAEE195 pKa = 4.52LVGGPVHH202 pKa = 5.9GTVSLAADD210 pKa = 4.25GSFTYY215 pKa = 10.34TPDD218 pKa = 3.51ADD220 pKa = 3.87YY221 pKa = 11.27SGPDD225 pKa = 3.06SFTYY229 pKa = 9.86KK230 pKa = 10.75AKK232 pKa = 10.84DD233 pKa = 3.36PGGLTSADD241 pKa = 3.56TTVSLTVTAVNDD253 pKa = 3.69APVAGDD259 pKa = 4.22DD260 pKa = 3.88SASTDD265 pKa = 3.13EE266 pKa = 4.31EE267 pKa = 4.49LAVDD271 pKa = 4.02VQVLEE276 pKa = 4.43NDD278 pKa = 3.55SDD280 pKa = 4.59PEE282 pKa = 4.3GSTLVVTGASGGTHH296 pKa = 5.94GTTAHH301 pKa = 7.0DD302 pKa = 4.41GDD304 pKa = 4.53SVTYY308 pKa = 9.64TPAPDD313 pKa = 3.61YY314 pKa = 11.01FGTDD318 pKa = 2.71SFTYY322 pKa = 10.22SVSDD326 pKa = 3.79GNGGTDD332 pKa = 3.07SATVTVTVSNVNDD345 pKa = 3.72APVAAGDD352 pKa = 3.78TGSTDD357 pKa = 3.17EE358 pKa = 5.6DD359 pKa = 3.79SALVVGASDD368 pKa = 3.96GLLANDD374 pKa = 4.26SDD376 pKa = 4.35VDD378 pKa = 4.02GDD380 pKa = 4.23SLTAALEE387 pKa = 4.61DD388 pKa = 4.62GPSHH392 pKa = 5.67GTVVLDD398 pKa = 3.63ADD400 pKa = 4.19GSYY403 pKa = 10.73SYY405 pKa = 10.93TPAADD410 pKa = 3.76YY411 pKa = 11.28YY412 pKa = 9.71GTDD415 pKa = 3.0SFTYY419 pKa = 9.72RR420 pKa = 11.84ANDD423 pKa = 3.79GAVSSGVATVTLTVAPVNDD442 pKa = 4.07APVADD447 pKa = 4.8DD448 pKa = 3.94EE449 pKa = 4.88SLTVAEE455 pKa = 5.11DD456 pKa = 3.49SQGSVDD462 pKa = 4.21VLLGDD467 pKa = 3.79TDD469 pKa = 4.1VEE471 pKa = 4.33GDD473 pKa = 3.86ALALTAVTEE482 pKa = 4.63GEE484 pKa = 4.26HH485 pKa = 5.71GTVVDD490 pKa = 4.31NGDD493 pKa = 3.38GTVTYY498 pKa = 10.0TPDD501 pKa = 3.4ADD503 pKa = 3.88YY504 pKa = 11.55AGADD508 pKa = 3.27SFTYY512 pKa = 8.18TVCDD516 pKa = 3.81DD517 pKa = 3.87HH518 pKa = 6.96ATTPTSSAGCDD529 pKa = 2.94TAMVEE534 pKa = 4.48VTVTPVNDD542 pKa = 3.82APVVTAGDD550 pKa = 3.48AASTDD555 pKa = 3.2EE556 pKa = 4.34GTAVTITATFTDD568 pKa = 3.13VDD570 pKa = 3.7RR571 pKa = 11.84AEE573 pKa = 4.73DD574 pKa = 3.55EE575 pKa = 4.74TYY577 pKa = 9.57TASIAWGDD585 pKa = 3.69GTTTAGTVAGNTVSGTHH602 pKa = 6.33TYY604 pKa = 10.99SDD606 pKa = 4.47DD607 pKa = 3.44EE608 pKa = 4.5AAPGDD613 pKa = 4.05DD614 pKa = 3.56VFPAVVTVTDD624 pKa = 3.54SGSTDD629 pKa = 3.32GVADD633 pKa = 4.41PLAGTATVAVTVANVAPTAATPALTVNPLTGLLSLSTSFSDD674 pKa = 4.0PAGSSDD680 pKa = 3.79TYY682 pKa = 11.03SGSFTVDD689 pKa = 2.75GDD691 pKa = 4.32TVPGTISGTTMSATTLLAPGCHH713 pKa = 5.02TVSATATVSDD723 pKa = 4.08EE724 pKa = 4.66DD725 pKa = 4.26GGSGTSGAGSVSQADD740 pKa = 4.02VYY742 pKa = 11.17AVSFQAPIRR751 pKa = 11.84DD752 pKa = 3.86DD753 pKa = 3.42VRR755 pKa = 11.84NVAKK759 pKa = 10.3YY760 pKa = 10.73GNVVPVKK767 pKa = 10.45VVITSSCTGLPVTTGPLAIQVVKK790 pKa = 11.11GNVTDD795 pKa = 5.03DD796 pKa = 3.63VTVDD800 pKa = 3.6DD801 pKa = 4.75SNAAVTEE808 pKa = 4.46SVSNADD814 pKa = 3.48SGTQMRR820 pKa = 11.84VADD823 pKa = 4.12GKK825 pKa = 11.03YY826 pKa = 9.81IYY828 pKa = 10.56NLSTKK833 pKa = 9.78GLTVGSDD840 pKa = 3.14YY841 pKa = 10.65TVRR844 pKa = 11.84IRR846 pKa = 11.84SGSTTGPVVLRR857 pKa = 11.84ALLSTKK863 pKa = 10.18KK864 pKa = 10.62
MM1 pKa = 7.97RR2 pKa = 11.84PPFLRR7 pKa = 11.84TTLAGMLVAATLGLAPAAFADD28 pKa = 3.71QTSRR32 pKa = 11.84DD33 pKa = 3.45GDD35 pKa = 3.76TAAVGRR41 pKa = 11.84NITYY45 pKa = 10.22GPGFRR50 pKa = 11.84ACTTRR55 pKa = 11.84GAQVSGAITVTYY67 pKa = 10.78NGGTHH72 pKa = 6.17LTPGEE77 pKa = 3.98ALVVTMSPPAGVTATAGATSVPATWNSNGQSFVIPLSTTVTTAAASGSVGFKK129 pKa = 10.68VVGASSGYY137 pKa = 10.57DD138 pKa = 3.22VTAGGSQYY146 pKa = 11.47DD147 pKa = 4.08VIVDD151 pKa = 4.28CPANTAPTGVADD163 pKa = 5.17SYY165 pKa = 11.67ATDD168 pKa = 3.42EE169 pKa = 4.32DD170 pKa = 4.29TAKK173 pKa = 10.6VVAAPGVLGNDD184 pKa = 3.6TDD186 pKa = 4.09PQGDD190 pKa = 3.94ALTAEE195 pKa = 4.52LVGGPVHH202 pKa = 5.9GTVSLAADD210 pKa = 4.25GSFTYY215 pKa = 10.34TPDD218 pKa = 3.51ADD220 pKa = 3.87YY221 pKa = 11.27SGPDD225 pKa = 3.06SFTYY229 pKa = 9.86KK230 pKa = 10.75AKK232 pKa = 10.84DD233 pKa = 3.36PGGLTSADD241 pKa = 3.56TTVSLTVTAVNDD253 pKa = 3.69APVAGDD259 pKa = 4.22DD260 pKa = 3.88SASTDD265 pKa = 3.13EE266 pKa = 4.31EE267 pKa = 4.49LAVDD271 pKa = 4.02VQVLEE276 pKa = 4.43NDD278 pKa = 3.55SDD280 pKa = 4.59PEE282 pKa = 4.3GSTLVVTGASGGTHH296 pKa = 5.94GTTAHH301 pKa = 7.0DD302 pKa = 4.41GDD304 pKa = 4.53SVTYY308 pKa = 9.64TPAPDD313 pKa = 3.61YY314 pKa = 11.01FGTDD318 pKa = 2.71SFTYY322 pKa = 10.22SVSDD326 pKa = 3.79GNGGTDD332 pKa = 3.07SATVTVTVSNVNDD345 pKa = 3.72APVAAGDD352 pKa = 3.78TGSTDD357 pKa = 3.17EE358 pKa = 5.6DD359 pKa = 3.79SALVVGASDD368 pKa = 3.96GLLANDD374 pKa = 4.26SDD376 pKa = 4.35VDD378 pKa = 4.02GDD380 pKa = 4.23SLTAALEE387 pKa = 4.61DD388 pKa = 4.62GPSHH392 pKa = 5.67GTVVLDD398 pKa = 3.63ADD400 pKa = 4.19GSYY403 pKa = 10.73SYY405 pKa = 10.93TPAADD410 pKa = 3.76YY411 pKa = 11.28YY412 pKa = 9.71GTDD415 pKa = 3.0SFTYY419 pKa = 9.72RR420 pKa = 11.84ANDD423 pKa = 3.79GAVSSGVATVTLTVAPVNDD442 pKa = 4.07APVADD447 pKa = 4.8DD448 pKa = 3.94EE449 pKa = 4.88SLTVAEE455 pKa = 5.11DD456 pKa = 3.49SQGSVDD462 pKa = 4.21VLLGDD467 pKa = 3.79TDD469 pKa = 4.1VEE471 pKa = 4.33GDD473 pKa = 3.86ALALTAVTEE482 pKa = 4.63GEE484 pKa = 4.26HH485 pKa = 5.71GTVVDD490 pKa = 4.31NGDD493 pKa = 3.38GTVTYY498 pKa = 10.0TPDD501 pKa = 3.4ADD503 pKa = 3.88YY504 pKa = 11.55AGADD508 pKa = 3.27SFTYY512 pKa = 8.18TVCDD516 pKa = 3.81DD517 pKa = 3.87HH518 pKa = 6.96ATTPTSSAGCDD529 pKa = 2.94TAMVEE534 pKa = 4.48VTVTPVNDD542 pKa = 3.82APVVTAGDD550 pKa = 3.48AASTDD555 pKa = 3.2EE556 pKa = 4.34GTAVTITATFTDD568 pKa = 3.13VDD570 pKa = 3.7RR571 pKa = 11.84AEE573 pKa = 4.73DD574 pKa = 3.55EE575 pKa = 4.74TYY577 pKa = 9.57TASIAWGDD585 pKa = 3.69GTTTAGTVAGNTVSGTHH602 pKa = 6.33TYY604 pKa = 10.99SDD606 pKa = 4.47DD607 pKa = 3.44EE608 pKa = 4.5AAPGDD613 pKa = 4.05DD614 pKa = 3.56VFPAVVTVTDD624 pKa = 3.54SGSTDD629 pKa = 3.32GVADD633 pKa = 4.41PLAGTATVAVTVANVAPTAATPALTVNPLTGLLSLSTSFSDD674 pKa = 4.0PAGSSDD680 pKa = 3.79TYY682 pKa = 11.03SGSFTVDD689 pKa = 2.75GDD691 pKa = 4.32TVPGTISGTTMSATTLLAPGCHH713 pKa = 5.02TVSATATVSDD723 pKa = 4.08EE724 pKa = 4.66DD725 pKa = 4.26GGSGTSGAGSVSQADD740 pKa = 4.02VYY742 pKa = 11.17AVSFQAPIRR751 pKa = 11.84DD752 pKa = 3.86DD753 pKa = 3.42VRR755 pKa = 11.84NVAKK759 pKa = 10.3YY760 pKa = 10.73GNVVPVKK767 pKa = 10.45VVITSSCTGLPVTTGPLAIQVVKK790 pKa = 11.11GNVTDD795 pKa = 5.03DD796 pKa = 3.63VTVDD800 pKa = 3.6DD801 pKa = 4.75SNAAVTEE808 pKa = 4.46SVSNADD814 pKa = 3.48SGTQMRR820 pKa = 11.84VADD823 pKa = 4.12GKK825 pKa = 11.03YY826 pKa = 9.81IYY828 pKa = 10.56NLSTKK833 pKa = 9.78GLTVGSDD840 pKa = 3.14YY841 pKa = 10.65TVRR844 pKa = 11.84IRR846 pKa = 11.84SGSTTGPVVLRR857 pKa = 11.84ALLSTKK863 pKa = 10.18KK864 pKa = 10.62
Molecular weight: 85.65 kDa
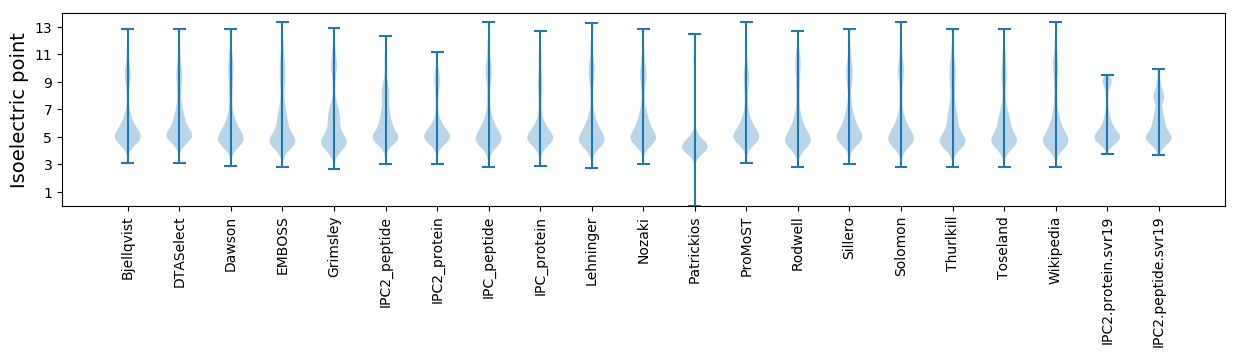
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2T5WB69|A0A2T5WB69_9ACTN DNA integrity scanning protein DisA OS=Nocardioides sp. CF204 OX=2135677 GN=disA PE=3 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.99KK16 pKa = 9.72HH17 pKa = 5.74RR18 pKa = 11.84KK19 pKa = 7.77LLKK22 pKa = 8.07RR23 pKa = 11.84TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 10.11LGKK33 pKa = 9.87
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.99KK16 pKa = 9.72HH17 pKa = 5.74RR18 pKa = 11.84KK19 pKa = 7.77LLKK22 pKa = 8.07RR23 pKa = 11.84TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 10.11LGKK33 pKa = 9.87
Molecular weight: 4.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1042830 |
29 |
2969 |
315.8 |
33.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.451 ± 0.066 | 0.736 ± 0.01 |
6.183 ± 0.038 | 5.899 ± 0.039 |
2.919 ± 0.022 | 9.123 ± 0.036 |
2.21 ± 0.019 | 3.707 ± 0.03 |
2.428 ± 0.036 | 10.272 ± 0.064 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.859 ± 0.018 | 1.886 ± 0.022 |
5.445 ± 0.034 | 2.794 ± 0.019 |
7.229 ± 0.045 | 5.1 ± 0.028 |
6.068 ± 0.041 | 9.193 ± 0.042 |
1.536 ± 0.018 | 1.962 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
