
Capybara microvirus Cap3_SP_389
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.17
Get precalculated fractions of proteins
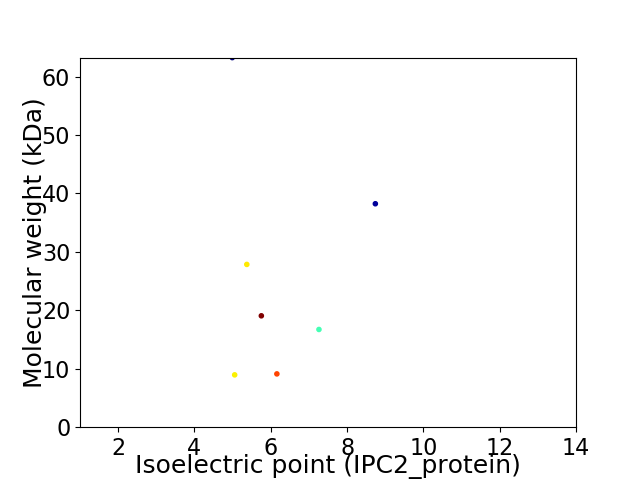
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
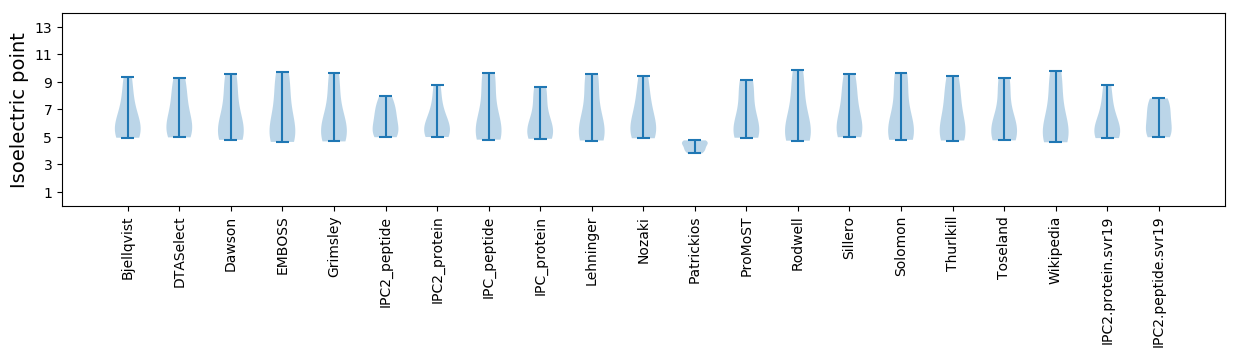
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4V1FVS8|A0A4V1FVS8_9VIRU Nonstructural protein OS=Capybara microvirus Cap3_SP_389 OX=2585441 PE=4 SV=1
MM1 pKa = 7.07NRR3 pKa = 11.84EE4 pKa = 4.14SNNYY8 pKa = 9.57FSDD11 pKa = 3.55LPYY14 pKa = 10.14IEE16 pKa = 4.71KK17 pKa = 10.15PRR19 pKa = 11.84SKK21 pKa = 10.33FSRR24 pKa = 11.84PQPHH28 pKa = 5.9ITTFKK33 pKa = 10.86NGDD36 pKa = 4.55LIPVYY41 pKa = 9.98IDD43 pKa = 3.17EE44 pKa = 4.58VLPGDD49 pKa = 4.78SMTIDD54 pKa = 3.28VNAVVRR60 pKa = 11.84MMTPIYY66 pKa = 10.2PVMDD70 pKa = 4.1NLILDD75 pKa = 3.89VMAFYY80 pKa = 10.88CPNRR84 pKa = 11.84LLWTHH89 pKa = 4.94WQEE92 pKa = 4.4FLGEE96 pKa = 4.19NKK98 pKa = 8.45LTAWEE103 pKa = 4.19QPVKK107 pKa = 11.25YY108 pKa = 9.57EE109 pKa = 3.87IPQIKK114 pKa = 9.94APEE117 pKa = 4.66GGWKK121 pKa = 10.34AGTIADD127 pKa = 3.79YY128 pKa = 11.15LGIPTYY134 pKa = 11.05VEE136 pKa = 4.0NLSVNAMPFRR146 pKa = 11.84AYY148 pKa = 10.96SLIWNEE154 pKa = 3.58FFRR157 pKa = 11.84DD158 pKa = 3.48EE159 pKa = 4.1NLKK162 pKa = 10.8DD163 pKa = 3.19RR164 pKa = 11.84VMINLDD170 pKa = 3.14DD171 pKa = 3.82ATRR174 pKa = 11.84VGSNGDD180 pKa = 3.63KK181 pKa = 11.19EE182 pKa = 4.25GDD184 pKa = 3.82TYY186 pKa = 11.14VTTAQLGAKK195 pKa = 8.06PLKK198 pKa = 9.47VAKK201 pKa = 8.98THH203 pKa = 7.73DD204 pKa = 4.56YY205 pKa = 7.87FTSALPNTQKK215 pKa = 11.19GEE217 pKa = 3.89AVLIPVISAGTGGLIPVTAGEE238 pKa = 3.97RR239 pKa = 11.84HH240 pKa = 5.91IEE242 pKa = 4.16NGSVTGNGLLWATNQSGLNNWWNVQYY268 pKa = 11.21DD269 pKa = 4.01GLSDD273 pKa = 3.57NKK275 pKa = 9.88TVSTAATGGTIHH287 pKa = 6.84NGPIVPANLYY297 pKa = 11.13ANLEE301 pKa = 4.31DD302 pKa = 4.04SQSLATIGTINEE314 pKa = 4.03LRR316 pKa = 11.84QSFAIQRR323 pKa = 11.84ILEE326 pKa = 4.04AMARR330 pKa = 11.84GGSRR334 pKa = 11.84YY335 pKa = 8.13TEE337 pKa = 3.59ILKK340 pKa = 10.33NIWGVTAPDD349 pKa = 3.5ARR351 pKa = 11.84LQRR354 pKa = 11.84PEE356 pKa = 3.86YY357 pKa = 10.42LGGEE361 pKa = 4.55RR362 pKa = 11.84IPINMDD368 pKa = 2.95QVLQTSASNGNNPLGDD384 pKa = 3.12TGAFSNTGFSTTLVNKK400 pKa = 10.26SFVEE404 pKa = 3.78HH405 pKa = 6.74GYY407 pKa = 10.71VIIVAAARR415 pKa = 11.84IRR417 pKa = 11.84NHH419 pKa = 6.76TYY421 pKa = 9.55QQGLEE426 pKa = 4.24KK427 pKa = 9.25MWSRR431 pKa = 11.84KK432 pKa = 7.37TMYY435 pKa = 10.35DD436 pKa = 3.01FYY438 pKa = 10.36MPQLAYY444 pKa = 10.66LSEE447 pKa = 3.91QAVLNKK453 pKa = 10.13EE454 pKa = 4.09IYY456 pKa = 9.85AQGNRR461 pKa = 11.84EE462 pKa = 4.0DD463 pKa = 4.32EE464 pKa = 4.39EE465 pKa = 4.74VFGYY469 pKa = 7.43QEE471 pKa = 3.15RR472 pKa = 11.84WAEE475 pKa = 3.82YY476 pKa = 8.27RR477 pKa = 11.84YY478 pKa = 10.64KK479 pKa = 10.45PGRR482 pKa = 11.84VSGAMRR488 pKa = 11.84SNYY491 pKa = 9.61PNGSLDD497 pKa = 3.19SWHH500 pKa = 6.46YY501 pKa = 11.42ADD503 pKa = 6.01DD504 pKa = 4.23YY505 pKa = 11.23VSKK508 pKa = 10.74PMLSSEE514 pKa = 4.41WIDD517 pKa = 3.43EE518 pKa = 4.28GTSEE522 pKa = 4.47VDD524 pKa = 3.01RR525 pKa = 11.84TIAIQSEE532 pKa = 4.48LEE534 pKa = 4.13DD535 pKa = 3.4QFLADD540 pKa = 5.28FLFINNSTRR549 pKa = 11.84TMPIYY554 pKa = 10.38SVPGLLDD561 pKa = 3.54HH562 pKa = 6.83YY563 pKa = 11.48
MM1 pKa = 7.07NRR3 pKa = 11.84EE4 pKa = 4.14SNNYY8 pKa = 9.57FSDD11 pKa = 3.55LPYY14 pKa = 10.14IEE16 pKa = 4.71KK17 pKa = 10.15PRR19 pKa = 11.84SKK21 pKa = 10.33FSRR24 pKa = 11.84PQPHH28 pKa = 5.9ITTFKK33 pKa = 10.86NGDD36 pKa = 4.55LIPVYY41 pKa = 9.98IDD43 pKa = 3.17EE44 pKa = 4.58VLPGDD49 pKa = 4.78SMTIDD54 pKa = 3.28VNAVVRR60 pKa = 11.84MMTPIYY66 pKa = 10.2PVMDD70 pKa = 4.1NLILDD75 pKa = 3.89VMAFYY80 pKa = 10.88CPNRR84 pKa = 11.84LLWTHH89 pKa = 4.94WQEE92 pKa = 4.4FLGEE96 pKa = 4.19NKK98 pKa = 8.45LTAWEE103 pKa = 4.19QPVKK107 pKa = 11.25YY108 pKa = 9.57EE109 pKa = 3.87IPQIKK114 pKa = 9.94APEE117 pKa = 4.66GGWKK121 pKa = 10.34AGTIADD127 pKa = 3.79YY128 pKa = 11.15LGIPTYY134 pKa = 11.05VEE136 pKa = 4.0NLSVNAMPFRR146 pKa = 11.84AYY148 pKa = 10.96SLIWNEE154 pKa = 3.58FFRR157 pKa = 11.84DD158 pKa = 3.48EE159 pKa = 4.1NLKK162 pKa = 10.8DD163 pKa = 3.19RR164 pKa = 11.84VMINLDD170 pKa = 3.14DD171 pKa = 3.82ATRR174 pKa = 11.84VGSNGDD180 pKa = 3.63KK181 pKa = 11.19EE182 pKa = 4.25GDD184 pKa = 3.82TYY186 pKa = 11.14VTTAQLGAKK195 pKa = 8.06PLKK198 pKa = 9.47VAKK201 pKa = 8.98THH203 pKa = 7.73DD204 pKa = 4.56YY205 pKa = 7.87FTSALPNTQKK215 pKa = 11.19GEE217 pKa = 3.89AVLIPVISAGTGGLIPVTAGEE238 pKa = 3.97RR239 pKa = 11.84HH240 pKa = 5.91IEE242 pKa = 4.16NGSVTGNGLLWATNQSGLNNWWNVQYY268 pKa = 11.21DD269 pKa = 4.01GLSDD273 pKa = 3.57NKK275 pKa = 9.88TVSTAATGGTIHH287 pKa = 6.84NGPIVPANLYY297 pKa = 11.13ANLEE301 pKa = 4.31DD302 pKa = 4.04SQSLATIGTINEE314 pKa = 4.03LRR316 pKa = 11.84QSFAIQRR323 pKa = 11.84ILEE326 pKa = 4.04AMARR330 pKa = 11.84GGSRR334 pKa = 11.84YY335 pKa = 8.13TEE337 pKa = 3.59ILKK340 pKa = 10.33NIWGVTAPDD349 pKa = 3.5ARR351 pKa = 11.84LQRR354 pKa = 11.84PEE356 pKa = 3.86YY357 pKa = 10.42LGGEE361 pKa = 4.55RR362 pKa = 11.84IPINMDD368 pKa = 2.95QVLQTSASNGNNPLGDD384 pKa = 3.12TGAFSNTGFSTTLVNKK400 pKa = 10.26SFVEE404 pKa = 3.78HH405 pKa = 6.74GYY407 pKa = 10.71VIIVAAARR415 pKa = 11.84IRR417 pKa = 11.84NHH419 pKa = 6.76TYY421 pKa = 9.55QQGLEE426 pKa = 4.24KK427 pKa = 9.25MWSRR431 pKa = 11.84KK432 pKa = 7.37TMYY435 pKa = 10.35DD436 pKa = 3.01FYY438 pKa = 10.36MPQLAYY444 pKa = 10.66LSEE447 pKa = 3.91QAVLNKK453 pKa = 10.13EE454 pKa = 4.09IYY456 pKa = 9.85AQGNRR461 pKa = 11.84EE462 pKa = 4.0DD463 pKa = 4.32EE464 pKa = 4.39EE465 pKa = 4.74VFGYY469 pKa = 7.43QEE471 pKa = 3.15RR472 pKa = 11.84WAEE475 pKa = 3.82YY476 pKa = 8.27RR477 pKa = 11.84YY478 pKa = 10.64KK479 pKa = 10.45PGRR482 pKa = 11.84VSGAMRR488 pKa = 11.84SNYY491 pKa = 9.61PNGSLDD497 pKa = 3.19SWHH500 pKa = 6.46YY501 pKa = 11.42ADD503 pKa = 6.01DD504 pKa = 4.23YY505 pKa = 11.23VSKK508 pKa = 10.74PMLSSEE514 pKa = 4.41WIDD517 pKa = 3.43EE518 pKa = 4.28GTSEE522 pKa = 4.47VDD524 pKa = 3.01RR525 pKa = 11.84TIAIQSEE532 pKa = 4.48LEE534 pKa = 4.13DD535 pKa = 3.4QFLADD540 pKa = 5.28FLFINNSTRR549 pKa = 11.84TMPIYY554 pKa = 10.38SVPGLLDD561 pKa = 3.54HH562 pKa = 6.83YY563 pKa = 11.48
Molecular weight: 63.26 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W8G2|A0A4P8W8G2_9VIRU Lysozyme OS=Capybara microvirus Cap3_SP_389 OX=2585441 PE=3 SV=1
MM1 pKa = 7.5SAIACIKK8 pKa = 9.61PIKK11 pKa = 9.94CWEE14 pKa = 4.04HH15 pKa = 5.92LKK17 pKa = 10.94KK18 pKa = 9.88KK19 pKa = 6.36TAKK22 pKa = 9.31GRR24 pKa = 11.84KK25 pKa = 8.83LISFKK30 pKa = 10.9QPTISDD36 pKa = 3.13IDD38 pKa = 3.48NWNEE42 pKa = 3.46IQIPCGQCIEE52 pKa = 4.09CRR54 pKa = 11.84LRR56 pKa = 11.84YY57 pKa = 8.9SRR59 pKa = 11.84HH60 pKa = 4.07WATRR64 pKa = 11.84LTLEE68 pKa = 4.42KK69 pKa = 10.11KK70 pKa = 7.22YY71 pKa = 10.78HH72 pKa = 6.08KK73 pKa = 10.05YY74 pKa = 10.85SYY76 pKa = 10.37FITLTYY82 pKa = 10.68DD83 pKa = 3.11EE84 pKa = 4.66EE85 pKa = 4.48HH86 pKa = 6.23LHH88 pKa = 5.74RR89 pKa = 11.84VEE91 pKa = 5.78NIDD94 pKa = 3.55TKK96 pKa = 10.4TGEE99 pKa = 4.05IVEE102 pKa = 4.68LSTLVKK108 pKa = 10.46KK109 pKa = 10.78DD110 pKa = 3.17MQDD113 pKa = 2.85FLKK116 pKa = 10.34RR117 pKa = 11.84YY118 pKa = 8.2RR119 pKa = 11.84YY120 pKa = 9.79YY121 pKa = 9.45YY122 pKa = 10.98GKK124 pKa = 10.09IKK126 pKa = 10.42FYY128 pKa = 10.75QCGEE132 pKa = 4.22YY133 pKa = 10.92GEE135 pKa = 4.37QTHH138 pKa = 6.38RR139 pKa = 11.84RR140 pKa = 11.84HH141 pKa = 5.26HH142 pKa = 6.08HH143 pKa = 5.6MIVFQEE149 pKa = 4.27RR150 pKa = 11.84PIEE153 pKa = 3.99PLEE156 pKa = 3.79FLKK159 pKa = 10.74LQKK162 pKa = 10.54DD163 pKa = 3.8NQPLYY168 pKa = 11.05TNLPLQKK175 pKa = 9.5IWGKK179 pKa = 10.76GMITIGEE186 pKa = 4.31VTPEE190 pKa = 3.63SRR192 pKa = 11.84GYY194 pKa = 8.08VARR197 pKa = 11.84YY198 pKa = 8.61VLKK201 pKa = 9.91KK202 pKa = 10.65QKK204 pKa = 10.78GEE206 pKa = 3.81SSNIYY211 pKa = 8.28NTLGILPEE219 pKa = 4.27YY220 pKa = 9.78VTMSNGIGMEE230 pKa = 3.95YY231 pKa = 10.62FKK233 pKa = 11.29DD234 pKa = 3.73NKK236 pKa = 9.2TKK238 pKa = 10.13IYY240 pKa = 10.18QYY242 pKa = 11.47DD243 pKa = 4.0SIILTGMHH251 pKa = 6.65GEE253 pKa = 4.07AKK255 pKa = 9.83KK256 pKa = 10.69VKK258 pKa = 9.4PPRR261 pKa = 11.84AFDD264 pKa = 3.49EE265 pKa = 4.41EE266 pKa = 4.3LAKK269 pKa = 10.98EE270 pKa = 4.01NLSEE274 pKa = 3.95ITSIKK279 pKa = 10.1DD280 pKa = 3.0KK281 pKa = 10.84RR282 pKa = 11.84RR283 pKa = 11.84AAAIRR288 pKa = 11.84GRR290 pKa = 11.84SVKK293 pKa = 9.63EE294 pKa = 3.63AQTEE298 pKa = 4.1NVNWAKK304 pKa = 10.75VEE306 pKa = 3.94SDD308 pKa = 3.55YY309 pKa = 11.55KK310 pKa = 10.2QARR313 pKa = 11.84QKK315 pKa = 10.5ILNKK319 pKa = 10.31RR320 pKa = 11.84EE321 pKa = 3.61VDD323 pKa = 3.71DD324 pKa = 4.16
MM1 pKa = 7.5SAIACIKK8 pKa = 9.61PIKK11 pKa = 9.94CWEE14 pKa = 4.04HH15 pKa = 5.92LKK17 pKa = 10.94KK18 pKa = 9.88KK19 pKa = 6.36TAKK22 pKa = 9.31GRR24 pKa = 11.84KK25 pKa = 8.83LISFKK30 pKa = 10.9QPTISDD36 pKa = 3.13IDD38 pKa = 3.48NWNEE42 pKa = 3.46IQIPCGQCIEE52 pKa = 4.09CRR54 pKa = 11.84LRR56 pKa = 11.84YY57 pKa = 8.9SRR59 pKa = 11.84HH60 pKa = 4.07WATRR64 pKa = 11.84LTLEE68 pKa = 4.42KK69 pKa = 10.11KK70 pKa = 7.22YY71 pKa = 10.78HH72 pKa = 6.08KK73 pKa = 10.05YY74 pKa = 10.85SYY76 pKa = 10.37FITLTYY82 pKa = 10.68DD83 pKa = 3.11EE84 pKa = 4.66EE85 pKa = 4.48HH86 pKa = 6.23LHH88 pKa = 5.74RR89 pKa = 11.84VEE91 pKa = 5.78NIDD94 pKa = 3.55TKK96 pKa = 10.4TGEE99 pKa = 4.05IVEE102 pKa = 4.68LSTLVKK108 pKa = 10.46KK109 pKa = 10.78DD110 pKa = 3.17MQDD113 pKa = 2.85FLKK116 pKa = 10.34RR117 pKa = 11.84YY118 pKa = 8.2RR119 pKa = 11.84YY120 pKa = 9.79YY121 pKa = 9.45YY122 pKa = 10.98GKK124 pKa = 10.09IKK126 pKa = 10.42FYY128 pKa = 10.75QCGEE132 pKa = 4.22YY133 pKa = 10.92GEE135 pKa = 4.37QTHH138 pKa = 6.38RR139 pKa = 11.84RR140 pKa = 11.84HH141 pKa = 5.26HH142 pKa = 6.08HH143 pKa = 5.6MIVFQEE149 pKa = 4.27RR150 pKa = 11.84PIEE153 pKa = 3.99PLEE156 pKa = 3.79FLKK159 pKa = 10.74LQKK162 pKa = 10.54DD163 pKa = 3.8NQPLYY168 pKa = 11.05TNLPLQKK175 pKa = 9.5IWGKK179 pKa = 10.76GMITIGEE186 pKa = 4.31VTPEE190 pKa = 3.63SRR192 pKa = 11.84GYY194 pKa = 8.08VARR197 pKa = 11.84YY198 pKa = 8.61VLKK201 pKa = 9.91KK202 pKa = 10.65QKK204 pKa = 10.78GEE206 pKa = 3.81SSNIYY211 pKa = 8.28NTLGILPEE219 pKa = 4.27YY220 pKa = 9.78VTMSNGIGMEE230 pKa = 3.95YY231 pKa = 10.62FKK233 pKa = 11.29DD234 pKa = 3.73NKK236 pKa = 9.2TKK238 pKa = 10.13IYY240 pKa = 10.18QYY242 pKa = 11.47DD243 pKa = 4.0SIILTGMHH251 pKa = 6.65GEE253 pKa = 4.07AKK255 pKa = 9.83KK256 pKa = 10.69VKK258 pKa = 9.4PPRR261 pKa = 11.84AFDD264 pKa = 3.49EE265 pKa = 4.41EE266 pKa = 4.3LAKK269 pKa = 10.98EE270 pKa = 4.01NLSEE274 pKa = 3.95ITSIKK279 pKa = 10.1DD280 pKa = 3.0KK281 pKa = 10.84RR282 pKa = 11.84RR283 pKa = 11.84AAAIRR288 pKa = 11.84GRR290 pKa = 11.84SVKK293 pKa = 9.63EE294 pKa = 3.63AQTEE298 pKa = 4.1NVNWAKK304 pKa = 10.75VEE306 pKa = 3.94SDD308 pKa = 3.55YY309 pKa = 11.55KK310 pKa = 10.2QARR313 pKa = 11.84QKK315 pKa = 10.5ILNKK319 pKa = 10.31RR320 pKa = 11.84EE321 pKa = 3.61VDD323 pKa = 3.71DD324 pKa = 4.16
Molecular weight: 38.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1605 |
74 |
563 |
229.3 |
26.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.854 ± 1.486 | 0.748 ± 0.334 |
5.483 ± 0.421 | 7.975 ± 0.86 |
3.427 ± 0.43 | 6.729 ± 0.502 |
1.62 ± 0.424 | 7.539 ± 0.74 |
7.477 ± 1.537 | 7.04 ± 0.647 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.617 ± 0.327 | 5.919 ± 0.785 |
3.614 ± 0.712 | 3.988 ± 0.214 |
5.234 ± 0.286 | 6.231 ± 0.969 |
6.355 ± 0.463 | 4.361 ± 0.585 |
1.807 ± 0.243 | 4.984 ± 0.425 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
