
Helicosporidium sp. ATCC 50920
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; core chlorophytes; Trebouxiophyceae; Chlorellales; Chlorellaceae; Helicosporidium; unclassified Helicosporidium
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
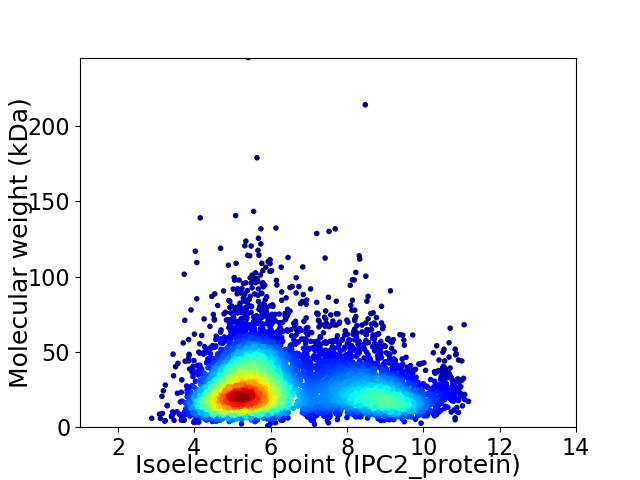
Virtual 2D-PAGE plot for 6029 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A059LJF9|A0A059LJF9_9CHLO Uncharacterized protein OS=Helicosporidium sp. ATCC 50920 OX=1291522 GN=H632_c1402p0 PE=4 SV=1
MM1 pKa = 7.87DD2 pKa = 5.16PAAEE6 pKa = 4.05HH7 pKa = 6.02FKK9 pKa = 11.08YY10 pKa = 10.04PVSIDD15 pKa = 2.88IRR17 pKa = 11.84EE18 pKa = 4.59LITLEE23 pKa = 5.53DD24 pKa = 3.63VMQEE28 pKa = 4.17LDD30 pKa = 3.57LGPNGGLLYY39 pKa = 10.74CMEE42 pKa = 4.23YY43 pKa = 9.78MEE45 pKa = 4.55EE46 pKa = 5.84HH47 pKa = 6.48LDD49 pKa = 3.31EE50 pKa = 4.48WLGQEE55 pKa = 4.4LEE57 pKa = 4.54SFGEE61 pKa = 3.95DD62 pKa = 3.46DD63 pKa = 5.49YY64 pKa = 12.21LLFDD68 pKa = 4.49CPGQIEE74 pKa = 5.2LYY76 pKa = 9.47SHH78 pKa = 6.64VNVFRR83 pKa = 11.84SFVDD87 pKa = 4.11FLRR90 pKa = 11.84KK91 pKa = 10.19DD92 pKa = 3.18GWQVCAVYY100 pKa = 10.83CMDD103 pKa = 3.58AQFVAEE109 pKa = 4.26APKK112 pKa = 10.65FIGGCLSALSAMVHH126 pKa = 6.5LEE128 pKa = 3.98LPHH131 pKa = 8.34INVLTKK137 pKa = 10.63ADD139 pKa = 5.04LIQNKK144 pKa = 8.85EE145 pKa = 3.95VLDD148 pKa = 4.87DD149 pKa = 3.75YY150 pKa = 11.22LFPSVEE156 pKa = 4.17DD157 pKa = 3.5MQHH160 pKa = 6.98RR161 pKa = 11.84LDD163 pKa = 3.88TTTAPHH169 pKa = 6.76LRR171 pKa = 11.84PLNAAMAQLVDD182 pKa = 3.55QFGLVSFVSLDD193 pKa = 3.28ITDD196 pKa = 4.01EE197 pKa = 4.06EE198 pKa = 5.07SIEE201 pKa = 4.01QLLLQVDD208 pKa = 3.72STIQYY213 pKa = 11.04GEE215 pKa = 4.08DD216 pKa = 3.09QEE218 pKa = 5.13VRR220 pKa = 11.84TKK222 pKa = 10.78EE223 pKa = 3.94YY224 pKa = 11.04GDD226 pKa = 4.08FPDD229 pKa = 4.49EE230 pKa = 4.06EE231 pKa = 4.44SAAAAGEE238 pKa = 4.11QEE240 pKa = 4.7LDD242 pKa = 3.63FF243 pKa = 6.37
MM1 pKa = 7.87DD2 pKa = 5.16PAAEE6 pKa = 4.05HH7 pKa = 6.02FKK9 pKa = 11.08YY10 pKa = 10.04PVSIDD15 pKa = 2.88IRR17 pKa = 11.84EE18 pKa = 4.59LITLEE23 pKa = 5.53DD24 pKa = 3.63VMQEE28 pKa = 4.17LDD30 pKa = 3.57LGPNGGLLYY39 pKa = 10.74CMEE42 pKa = 4.23YY43 pKa = 9.78MEE45 pKa = 4.55EE46 pKa = 5.84HH47 pKa = 6.48LDD49 pKa = 3.31EE50 pKa = 4.48WLGQEE55 pKa = 4.4LEE57 pKa = 4.54SFGEE61 pKa = 3.95DD62 pKa = 3.46DD63 pKa = 5.49YY64 pKa = 12.21LLFDD68 pKa = 4.49CPGQIEE74 pKa = 5.2LYY76 pKa = 9.47SHH78 pKa = 6.64VNVFRR83 pKa = 11.84SFVDD87 pKa = 4.11FLRR90 pKa = 11.84KK91 pKa = 10.19DD92 pKa = 3.18GWQVCAVYY100 pKa = 10.83CMDD103 pKa = 3.58AQFVAEE109 pKa = 4.26APKK112 pKa = 10.65FIGGCLSALSAMVHH126 pKa = 6.5LEE128 pKa = 3.98LPHH131 pKa = 8.34INVLTKK137 pKa = 10.63ADD139 pKa = 5.04LIQNKK144 pKa = 8.85EE145 pKa = 3.95VLDD148 pKa = 4.87DD149 pKa = 3.75YY150 pKa = 11.22LFPSVEE156 pKa = 4.17DD157 pKa = 3.5MQHH160 pKa = 6.98RR161 pKa = 11.84LDD163 pKa = 3.88TTTAPHH169 pKa = 6.76LRR171 pKa = 11.84PLNAAMAQLVDD182 pKa = 3.55QFGLVSFVSLDD193 pKa = 3.28ITDD196 pKa = 4.01EE197 pKa = 4.06EE198 pKa = 5.07SIEE201 pKa = 4.01QLLLQVDD208 pKa = 3.72STIQYY213 pKa = 11.04GEE215 pKa = 4.08DD216 pKa = 3.09QEE218 pKa = 5.13VRR220 pKa = 11.84TKK222 pKa = 10.78EE223 pKa = 3.94YY224 pKa = 11.04GDD226 pKa = 4.08FPDD229 pKa = 4.49EE230 pKa = 4.06EE231 pKa = 4.44SAAAAGEE238 pKa = 4.11QEE240 pKa = 4.7LDD242 pKa = 3.63FF243 pKa = 6.37
Molecular weight: 27.59 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A059LAK7|A0A059LAK7_9CHLO Glucose inhibited division protein A (Fragment) OS=Helicosporidium sp. ATCC 50920 OX=1291522 GN=H632_c5143p0 PE=3 SV=1
RR1 pKa = 7.05QALPRR6 pKa = 11.84ARR8 pKa = 11.84VGPLGRR14 pKa = 11.84LLGKK18 pKa = 10.05KK19 pKa = 8.89RR20 pKa = 11.84ASGRR24 pKa = 11.84GVDD27 pKa = 4.05GGAGGAARR35 pKa = 11.84PAPGRR40 pKa = 11.84GGGPGGARR48 pKa = 11.84AGVVLRR54 pKa = 11.84ARR56 pKa = 11.84LRR58 pKa = 11.84VEE60 pKa = 3.9RR61 pKa = 11.84RR62 pKa = 11.84AAQAGGRR69 pKa = 11.84GQLRR73 pKa = 11.84GGSRR77 pKa = 11.84FANGLGAAPRR87 pKa = 11.84APALLGSPSSRR98 pKa = 11.84ARR100 pKa = 11.84VAGGEE105 pKa = 3.95PGRR108 pKa = 11.84PGALRR113 pKa = 11.84RR114 pKa = 11.84ALWPGRR120 pKa = 11.84PGRR123 pKa = 11.84GRR125 pKa = 11.84LSQPLRR131 pKa = 11.84PLGRR135 pKa = 11.84RR136 pKa = 11.84WQGAARR142 pKa = 11.84PARR145 pKa = 11.84RR146 pKa = 11.84ARR148 pKa = 11.84RR149 pKa = 11.84ARR151 pKa = 11.84GRR153 pKa = 11.84PRR155 pKa = 11.84PAARR159 pKa = 11.84ARR161 pKa = 11.84GRR163 pKa = 11.84LASLL167 pKa = 3.83
RR1 pKa = 7.05QALPRR6 pKa = 11.84ARR8 pKa = 11.84VGPLGRR14 pKa = 11.84LLGKK18 pKa = 10.05KK19 pKa = 8.89RR20 pKa = 11.84ASGRR24 pKa = 11.84GVDD27 pKa = 4.05GGAGGAARR35 pKa = 11.84PAPGRR40 pKa = 11.84GGGPGGARR48 pKa = 11.84AGVVLRR54 pKa = 11.84ARR56 pKa = 11.84LRR58 pKa = 11.84VEE60 pKa = 3.9RR61 pKa = 11.84RR62 pKa = 11.84AAQAGGRR69 pKa = 11.84GQLRR73 pKa = 11.84GGSRR77 pKa = 11.84FANGLGAAPRR87 pKa = 11.84APALLGSPSSRR98 pKa = 11.84ARR100 pKa = 11.84VAGGEE105 pKa = 3.95PGRR108 pKa = 11.84PGALRR113 pKa = 11.84RR114 pKa = 11.84ALWPGRR120 pKa = 11.84PGRR123 pKa = 11.84GRR125 pKa = 11.84LSQPLRR131 pKa = 11.84PLGRR135 pKa = 11.84RR136 pKa = 11.84WQGAARR142 pKa = 11.84PARR145 pKa = 11.84RR146 pKa = 11.84ARR148 pKa = 11.84RR149 pKa = 11.84ARR151 pKa = 11.84GRR153 pKa = 11.84PRR155 pKa = 11.84PAARR159 pKa = 11.84ARR161 pKa = 11.84GRR163 pKa = 11.84LASLL167 pKa = 3.83
Molecular weight: 17.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1647305 |
8 |
2258 |
273.2 |
29.61 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.754 ± 0.05 | 1.548 ± 0.016 |
5.203 ± 0.023 | 6.626 ± 0.042 |
3.157 ± 0.018 | 8.295 ± 0.039 |
2.23 ± 0.016 | 2.802 ± 0.021 |
3.395 ± 0.03 | 10.198 ± 0.036 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.992 ± 0.015 | 2.151 ± 0.017 |
5.997 ± 0.027 | 3.598 ± 0.019 |
7.749 ± 0.045 | 7.149 ± 0.034 |
4.316 ± 0.022 | 7.202 ± 0.029 |
1.455 ± 0.013 | 2.183 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
