
Roseobacter phage DSS3P8
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 5.87
Get precalculated fractions of proteins
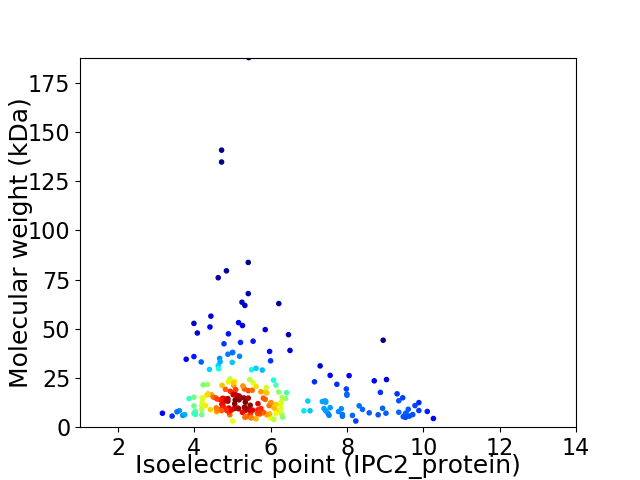
Virtual 2D-PAGE plot for 229 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1B0UXN0|A0A1B0UXN0_9CAUD Uncharacterized protein OS=Roseobacter phage DSS3P8 OX=1735562 GN=DSS3P8_124 PE=4 SV=1
MM1 pKa = 7.55FFPNNVSDD9 pKa = 3.66QSISVFFAGGMRR21 pKa = 11.84SIPASDD27 pKa = 3.58PGFAEE32 pKa = 4.9LSAHH36 pKa = 6.62LALGEE41 pKa = 4.01HH42 pKa = 7.4DD43 pKa = 4.25YY44 pKa = 10.46EE45 pKa = 4.52TVEE48 pKa = 4.7RR49 pKa = 11.84LVDD52 pKa = 3.77KK53 pKa = 10.68PKK55 pKa = 10.27MIARR59 pKa = 11.84LSEE62 pKa = 3.79GLVQVIGSTVYY73 pKa = 10.71YY74 pKa = 10.01KK75 pKa = 9.68GTPAHH80 pKa = 5.58STIAYY85 pKa = 9.04RR86 pKa = 11.84LLDD89 pKa = 3.8ILNAGQNAQVWARR102 pKa = 11.84FLEE105 pKa = 4.12RR106 pKa = 11.84VMANPSARR114 pKa = 11.84SRR116 pKa = 11.84QCLYY120 pKa = 10.95EE121 pKa = 5.68FLDD124 pKa = 3.39AWKK127 pKa = 10.72APLTEE132 pKa = 5.12DD133 pKa = 3.67GCFITFKK140 pKa = 10.43RR141 pKa = 11.84VRR143 pKa = 11.84ADD145 pKa = 3.05YY146 pKa = 11.07RR147 pKa = 11.84DD148 pKa = 3.29IHH150 pKa = 7.15SGKK153 pKa = 9.9FDD155 pKa = 3.69NSPGQTVEE163 pKa = 3.84VSRR166 pKa = 11.84DD167 pKa = 3.63EE168 pKa = 5.07VDD170 pKa = 4.36DD171 pKa = 6.17DD172 pKa = 4.3PDD174 pKa = 3.71EE175 pKa = 4.51TCSYY179 pKa = 9.18GLHH182 pKa = 5.65VAATSYY188 pKa = 10.71LGHH191 pKa = 7.05HH192 pKa = 6.02YY193 pKa = 10.6VSSPSYY199 pKa = 9.52NTIACKK205 pKa = 10.15VDD207 pKa = 3.37PADD210 pKa = 3.62VVAVPRR216 pKa = 11.84DD217 pKa = 3.51YY218 pKa = 11.51GSAKK222 pKa = 9.47MRR224 pKa = 11.84VCKK227 pKa = 10.59YY228 pKa = 10.36VVLGDD233 pKa = 4.06AEE235 pKa = 4.06EE236 pKa = 4.33SFYY239 pKa = 11.77NNAEE243 pKa = 3.97SSPIVPLSMTTSDD256 pKa = 4.19DD257 pKa = 3.67MAPEE261 pKa = 4.49VYY263 pKa = 10.62VDD265 pKa = 5.89DD266 pKa = 6.0DD267 pKa = 6.07FEE269 pKa = 7.2DD270 pKa = 3.78EE271 pKa = 4.52DD272 pKa = 4.68NEE274 pKa = 5.22DD275 pKa = 5.2RR276 pKa = 11.84ICPDD280 pKa = 3.35CGDD283 pKa = 4.62YY284 pKa = 10.88KK285 pKa = 11.28ADD287 pKa = 3.27WQAICEE293 pKa = 4.22DD294 pKa = 4.36CEE296 pKa = 3.96DD297 pKa = 3.87ARR299 pKa = 11.84NEE301 pKa = 4.11EE302 pKa = 4.14EE303 pKa = 5.44DD304 pKa = 3.7IPYY307 pKa = 10.44CGNCGNVEE315 pKa = 3.87VDD317 pKa = 5.39EE318 pKa = 5.33EE319 pKa = 5.35DD320 pKa = 4.4DD321 pKa = 3.87LCDD324 pKa = 3.18RR325 pKa = 11.84CEE327 pKa = 4.15QEE329 pKa = 5.48EE330 pKa = 4.04HH331 pKa = 7.37AEE333 pKa = 4.0WQEE336 pKa = 3.7QWDD339 pKa = 3.59ADD341 pKa = 3.48ARR343 pKa = 11.84AMEE346 pKa = 4.6EE347 pKa = 4.01EE348 pKa = 4.35DD349 pKa = 4.85CIAEE353 pKa = 4.32TGCTLNGVPVSDD365 pKa = 5.51DD366 pKa = 3.43PSDD369 pKa = 3.97YY370 pKa = 11.53DD371 pKa = 5.77DD372 pKa = 5.4IPDD375 pKa = 3.89TEE377 pKa = 4.13ASEE380 pKa = 4.22MAFMRR385 pKa = 11.84GGVTYY390 pKa = 9.87QASEE394 pKa = 4.01IMVGIANHH402 pKa = 5.15GQRR405 pKa = 11.84GYY407 pKa = 11.49SRR409 pKa = 11.84MTGIPRR415 pKa = 11.84TTLQDD420 pKa = 2.84WTTKK424 pKa = 10.23IEE426 pKa = 3.99ACEE429 pKa = 3.77
MM1 pKa = 7.55FFPNNVSDD9 pKa = 3.66QSISVFFAGGMRR21 pKa = 11.84SIPASDD27 pKa = 3.58PGFAEE32 pKa = 4.9LSAHH36 pKa = 6.62LALGEE41 pKa = 4.01HH42 pKa = 7.4DD43 pKa = 4.25YY44 pKa = 10.46EE45 pKa = 4.52TVEE48 pKa = 4.7RR49 pKa = 11.84LVDD52 pKa = 3.77KK53 pKa = 10.68PKK55 pKa = 10.27MIARR59 pKa = 11.84LSEE62 pKa = 3.79GLVQVIGSTVYY73 pKa = 10.71YY74 pKa = 10.01KK75 pKa = 9.68GTPAHH80 pKa = 5.58STIAYY85 pKa = 9.04RR86 pKa = 11.84LLDD89 pKa = 3.8ILNAGQNAQVWARR102 pKa = 11.84FLEE105 pKa = 4.12RR106 pKa = 11.84VMANPSARR114 pKa = 11.84SRR116 pKa = 11.84QCLYY120 pKa = 10.95EE121 pKa = 5.68FLDD124 pKa = 3.39AWKK127 pKa = 10.72APLTEE132 pKa = 5.12DD133 pKa = 3.67GCFITFKK140 pKa = 10.43RR141 pKa = 11.84VRR143 pKa = 11.84ADD145 pKa = 3.05YY146 pKa = 11.07RR147 pKa = 11.84DD148 pKa = 3.29IHH150 pKa = 7.15SGKK153 pKa = 9.9FDD155 pKa = 3.69NSPGQTVEE163 pKa = 3.84VSRR166 pKa = 11.84DD167 pKa = 3.63EE168 pKa = 5.07VDD170 pKa = 4.36DD171 pKa = 6.17DD172 pKa = 4.3PDD174 pKa = 3.71EE175 pKa = 4.51TCSYY179 pKa = 9.18GLHH182 pKa = 5.65VAATSYY188 pKa = 10.71LGHH191 pKa = 7.05HH192 pKa = 6.02YY193 pKa = 10.6VSSPSYY199 pKa = 9.52NTIACKK205 pKa = 10.15VDD207 pKa = 3.37PADD210 pKa = 3.62VVAVPRR216 pKa = 11.84DD217 pKa = 3.51YY218 pKa = 11.51GSAKK222 pKa = 9.47MRR224 pKa = 11.84VCKK227 pKa = 10.59YY228 pKa = 10.36VVLGDD233 pKa = 4.06AEE235 pKa = 4.06EE236 pKa = 4.33SFYY239 pKa = 11.77NNAEE243 pKa = 3.97SSPIVPLSMTTSDD256 pKa = 4.19DD257 pKa = 3.67MAPEE261 pKa = 4.49VYY263 pKa = 10.62VDD265 pKa = 5.89DD266 pKa = 6.0DD267 pKa = 6.07FEE269 pKa = 7.2DD270 pKa = 3.78EE271 pKa = 4.52DD272 pKa = 4.68NEE274 pKa = 5.22DD275 pKa = 5.2RR276 pKa = 11.84ICPDD280 pKa = 3.35CGDD283 pKa = 4.62YY284 pKa = 10.88KK285 pKa = 11.28ADD287 pKa = 3.27WQAICEE293 pKa = 4.22DD294 pKa = 4.36CEE296 pKa = 3.96DD297 pKa = 3.87ARR299 pKa = 11.84NEE301 pKa = 4.11EE302 pKa = 4.14EE303 pKa = 5.44DD304 pKa = 3.7IPYY307 pKa = 10.44CGNCGNVEE315 pKa = 3.87VDD317 pKa = 5.39EE318 pKa = 5.33EE319 pKa = 5.35DD320 pKa = 4.4DD321 pKa = 3.87LCDD324 pKa = 3.18RR325 pKa = 11.84CEE327 pKa = 4.15QEE329 pKa = 5.48EE330 pKa = 4.04HH331 pKa = 7.37AEE333 pKa = 4.0WQEE336 pKa = 3.7QWDD339 pKa = 3.59ADD341 pKa = 3.48ARR343 pKa = 11.84AMEE346 pKa = 4.6EE347 pKa = 4.01EE348 pKa = 4.35DD349 pKa = 4.85CIAEE353 pKa = 4.32TGCTLNGVPVSDD365 pKa = 5.51DD366 pKa = 3.43PSDD369 pKa = 3.97YY370 pKa = 11.53DD371 pKa = 5.77DD372 pKa = 5.4IPDD375 pKa = 3.89TEE377 pKa = 4.13ASEE380 pKa = 4.22MAFMRR385 pKa = 11.84GGVTYY390 pKa = 9.87QASEE394 pKa = 4.01IMVGIANHH402 pKa = 5.15GQRR405 pKa = 11.84GYY407 pKa = 11.49SRR409 pKa = 11.84MTGIPRR415 pKa = 11.84TTLQDD420 pKa = 2.84WTTKK424 pKa = 10.23IEE426 pKa = 3.99ACEE429 pKa = 3.77
Molecular weight: 47.85 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B0V1V5|A0A1B0V1V5_9CAUD Uncharacterized protein OS=Roseobacter phage DSS3P8 OX=1735562 GN=DSS3P8_003 PE=4 SV=1
MM1 pKa = 7.69GSPVHH6 pKa = 6.92ACQCKK11 pKa = 9.2DD12 pKa = 3.28CKK14 pKa = 9.97KK15 pKa = 10.46ARR17 pKa = 11.84RR18 pKa = 11.84KK19 pKa = 10.49GLVLGSRR26 pKa = 11.84RR27 pKa = 11.84MGFWCARR34 pKa = 11.84IIGYY38 pKa = 8.95GVEE41 pKa = 4.32YY42 pKa = 10.91YY43 pKa = 10.48LGSVSAYY50 pKa = 9.88VPHH53 pKa = 7.19PRR55 pKa = 11.84LAKK58 pKa = 9.7GKK60 pKa = 9.71RR61 pKa = 11.84LRR63 pKa = 11.84RR64 pKa = 11.84EE65 pKa = 3.68QNKK68 pKa = 9.01LQRR71 pKa = 11.84EE72 pKa = 4.02IRR74 pKa = 11.84TNMEE78 pKa = 3.89DD79 pKa = 3.06
MM1 pKa = 7.69GSPVHH6 pKa = 6.92ACQCKK11 pKa = 9.2DD12 pKa = 3.28CKK14 pKa = 9.97KK15 pKa = 10.46ARR17 pKa = 11.84RR18 pKa = 11.84KK19 pKa = 10.49GLVLGSRR26 pKa = 11.84RR27 pKa = 11.84MGFWCARR34 pKa = 11.84IIGYY38 pKa = 8.95GVEE41 pKa = 4.32YY42 pKa = 10.91YY43 pKa = 10.48LGSVSAYY50 pKa = 9.88VPHH53 pKa = 7.19PRR55 pKa = 11.84LAKK58 pKa = 9.7GKK60 pKa = 9.71RR61 pKa = 11.84LRR63 pKa = 11.84RR64 pKa = 11.84EE65 pKa = 3.68QNKK68 pKa = 9.01LQRR71 pKa = 11.84EE72 pKa = 4.02IRR74 pKa = 11.84TNMEE78 pKa = 3.89DD79 pKa = 3.06
Molecular weight: 9.13 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
40708 |
28 |
1825 |
177.8 |
19.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.634 ± 0.417 | 1.098 ± 0.088 |
6.755 ± 0.156 | 6.846 ± 0.197 |
3.913 ± 0.109 | 7.753 ± 0.218 |
2.076 ± 0.116 | 5.431 ± 0.139 |
4.532 ± 0.199 | 7.335 ± 0.147 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.225 ± 0.096 | 3.562 ± 0.113 |
4.267 ± 0.181 | 2.867 ± 0.16 |
5.974 ± 0.138 | 5.64 ± 0.243 |
6.281 ± 0.167 | 6.994 ± 0.13 |
1.619 ± 0.078 | 3.196 ± 0.141 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
