
Stackebrandtia endophytica
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Glycomycetales; Glycomycetaceae; Stackebrandtia
Average proteome isoelectric point is 6.02
Get precalculated fractions of proteins
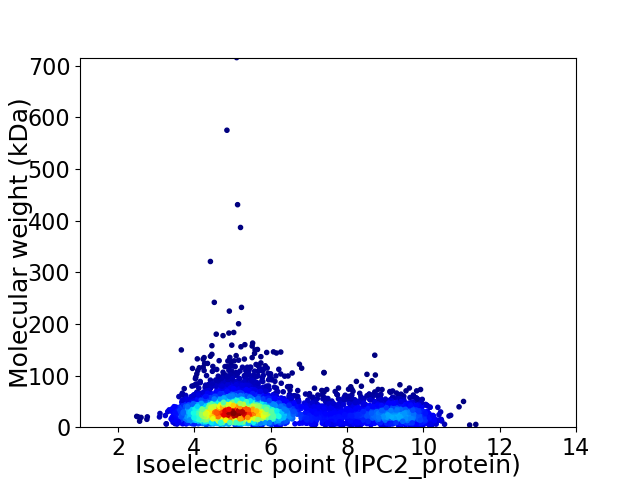
Virtual 2D-PAGE plot for 5197 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A543AST5|A0A543AST5_9ACTN (R)-citramalate synthase OS=Stackebrandtia endophytica OX=1496996 GN=FB566_1164 PE=3 SV=1
MM1 pKa = 6.99TPRR4 pKa = 11.84VRR6 pKa = 11.84GVLVGAVSLAVMLTGCASEE25 pKa = 4.34SAPLVQSDD33 pKa = 4.05EE34 pKa = 4.85DD35 pKa = 4.23DD36 pKa = 3.83VAVDD40 pKa = 4.27PNSGDD45 pKa = 3.49LSAGEE50 pKa = 4.55SDD52 pKa = 3.9PVTDD56 pKa = 3.79PFYY59 pKa = 10.73PEE61 pKa = 4.92FGNADD66 pKa = 3.57IDD68 pKa = 4.07VLHH71 pKa = 6.92YY72 pKa = 11.06DD73 pKa = 4.06LDD75 pKa = 4.09LSYY78 pKa = 11.1EE79 pKa = 4.02PSNQVLTGTATLTIRR94 pKa = 11.84PVVDD98 pKa = 3.28AAEE101 pKa = 4.27VAVDD105 pKa = 3.77LAKK108 pKa = 10.6PLMVSKK114 pKa = 9.67VTVDD118 pKa = 3.33DD119 pKa = 4.33VEE121 pKa = 4.43ATFQHH126 pKa = 5.55EE127 pKa = 4.37QYY129 pKa = 11.42DD130 pKa = 4.25LVIDD134 pKa = 3.94TAVTADD140 pKa = 3.56QPVVARR146 pKa = 11.84IEE148 pKa = 4.24YY149 pKa = 10.21SGVPQTVPAPAKK161 pKa = 10.25RR162 pKa = 11.84GDD164 pKa = 3.22MSMGIGASTNAVTGALWSFQEE185 pKa = 4.21PFGALTWYY193 pKa = 9.14PVNDD197 pKa = 4.12HH198 pKa = 7.12PSDD201 pKa = 3.47EE202 pKa = 4.27ALYY205 pKa = 10.38DD206 pKa = 3.51IAITVPDD213 pKa = 5.3GYY215 pKa = 11.23SGVASGKK222 pKa = 10.12FVGQEE227 pKa = 3.13GDD229 pKa = 3.63TFHH232 pKa = 6.57WTSSDD237 pKa = 3.21PVASYY242 pKa = 8.98VTTIAVDD249 pKa = 3.54QYY251 pKa = 11.54EE252 pKa = 4.48LTEE255 pKa = 3.98LTGPNGLPITLWTLPEE271 pKa = 4.06YY272 pKa = 11.03SEE274 pKa = 4.16FMPVLEE280 pKa = 5.02RR281 pKa = 11.84MPDD284 pKa = 3.35MIEE287 pKa = 3.89WLEE290 pKa = 4.02SRR292 pKa = 11.84YY293 pKa = 10.51GPYY296 pKa = 10.41PFDD299 pKa = 3.3SAGVVLVGGEE309 pKa = 4.27SAMEE313 pKa = 4.06TQEE316 pKa = 4.45MITFSGDD323 pKa = 3.78FINLPGVDD331 pKa = 3.78ADD333 pKa = 4.0YY334 pKa = 10.8VAGVVVHH341 pKa = 6.5EE342 pKa = 5.48LAHH345 pKa = 5.21QWFGNAVSPRR355 pKa = 11.84DD356 pKa = 3.85WSNLWLNEE364 pKa = 3.5GAATYY369 pKa = 9.87IEE371 pKa = 4.4QMWNVDD377 pKa = 2.88NGYY380 pKa = 6.78TTEE383 pKa = 4.12SSVVSGWEE391 pKa = 3.54RR392 pKa = 11.84DD393 pKa = 3.6DD394 pKa = 4.1EE395 pKa = 4.59YY396 pKa = 11.71LRR398 pKa = 11.84MAYY401 pKa = 9.64GPPGAADD408 pKa = 3.63PDD410 pKa = 3.94AFASSNSYY418 pKa = 9.95VCPALMLHH426 pKa = 7.03RR427 pKa = 11.84MRR429 pKa = 11.84DD430 pKa = 3.72LLGGADD436 pKa = 4.66KK437 pKa = 11.15VDD439 pKa = 3.66EE440 pKa = 5.05LLAAWVVEE448 pKa = 3.7FDD450 pKa = 3.48NRR452 pKa = 11.84SVEE455 pKa = 3.95RR456 pKa = 11.84DD457 pKa = 3.42DD458 pKa = 5.82FVAFVNEE465 pKa = 4.35YY466 pKa = 10.2TGTDD470 pKa = 2.98HH471 pKa = 7.29TAFIDD476 pKa = 3.44EE477 pKa = 4.45WLDD480 pKa = 3.77SEE482 pKa = 4.71TTPEE486 pKa = 3.7
MM1 pKa = 6.99TPRR4 pKa = 11.84VRR6 pKa = 11.84GVLVGAVSLAVMLTGCASEE25 pKa = 4.34SAPLVQSDD33 pKa = 4.05EE34 pKa = 4.85DD35 pKa = 4.23DD36 pKa = 3.83VAVDD40 pKa = 4.27PNSGDD45 pKa = 3.49LSAGEE50 pKa = 4.55SDD52 pKa = 3.9PVTDD56 pKa = 3.79PFYY59 pKa = 10.73PEE61 pKa = 4.92FGNADD66 pKa = 3.57IDD68 pKa = 4.07VLHH71 pKa = 6.92YY72 pKa = 11.06DD73 pKa = 4.06LDD75 pKa = 4.09LSYY78 pKa = 11.1EE79 pKa = 4.02PSNQVLTGTATLTIRR94 pKa = 11.84PVVDD98 pKa = 3.28AAEE101 pKa = 4.27VAVDD105 pKa = 3.77LAKK108 pKa = 10.6PLMVSKK114 pKa = 9.67VTVDD118 pKa = 3.33DD119 pKa = 4.33VEE121 pKa = 4.43ATFQHH126 pKa = 5.55EE127 pKa = 4.37QYY129 pKa = 11.42DD130 pKa = 4.25LVIDD134 pKa = 3.94TAVTADD140 pKa = 3.56QPVVARR146 pKa = 11.84IEE148 pKa = 4.24YY149 pKa = 10.21SGVPQTVPAPAKK161 pKa = 10.25RR162 pKa = 11.84GDD164 pKa = 3.22MSMGIGASTNAVTGALWSFQEE185 pKa = 4.21PFGALTWYY193 pKa = 9.14PVNDD197 pKa = 4.12HH198 pKa = 7.12PSDD201 pKa = 3.47EE202 pKa = 4.27ALYY205 pKa = 10.38DD206 pKa = 3.51IAITVPDD213 pKa = 5.3GYY215 pKa = 11.23SGVASGKK222 pKa = 10.12FVGQEE227 pKa = 3.13GDD229 pKa = 3.63TFHH232 pKa = 6.57WTSSDD237 pKa = 3.21PVASYY242 pKa = 8.98VTTIAVDD249 pKa = 3.54QYY251 pKa = 11.54EE252 pKa = 4.48LTEE255 pKa = 3.98LTGPNGLPITLWTLPEE271 pKa = 4.06YY272 pKa = 11.03SEE274 pKa = 4.16FMPVLEE280 pKa = 5.02RR281 pKa = 11.84MPDD284 pKa = 3.35MIEE287 pKa = 3.89WLEE290 pKa = 4.02SRR292 pKa = 11.84YY293 pKa = 10.51GPYY296 pKa = 10.41PFDD299 pKa = 3.3SAGVVLVGGEE309 pKa = 4.27SAMEE313 pKa = 4.06TQEE316 pKa = 4.45MITFSGDD323 pKa = 3.78FINLPGVDD331 pKa = 3.78ADD333 pKa = 4.0YY334 pKa = 10.8VAGVVVHH341 pKa = 6.5EE342 pKa = 5.48LAHH345 pKa = 5.21QWFGNAVSPRR355 pKa = 11.84DD356 pKa = 3.85WSNLWLNEE364 pKa = 3.5GAATYY369 pKa = 9.87IEE371 pKa = 4.4QMWNVDD377 pKa = 2.88NGYY380 pKa = 6.78TTEE383 pKa = 4.12SSVVSGWEE391 pKa = 3.54RR392 pKa = 11.84DD393 pKa = 3.6DD394 pKa = 4.1EE395 pKa = 4.59YY396 pKa = 11.71LRR398 pKa = 11.84MAYY401 pKa = 9.64GPPGAADD408 pKa = 3.63PDD410 pKa = 3.94AFASSNSYY418 pKa = 9.95VCPALMLHH426 pKa = 7.03RR427 pKa = 11.84MRR429 pKa = 11.84DD430 pKa = 3.72LLGGADD436 pKa = 4.66KK437 pKa = 11.15VDD439 pKa = 3.66EE440 pKa = 5.05LLAAWVVEE448 pKa = 3.7FDD450 pKa = 3.48NRR452 pKa = 11.84SVEE455 pKa = 3.95RR456 pKa = 11.84DD457 pKa = 3.42DD458 pKa = 5.82FVAFVNEE465 pKa = 4.35YY466 pKa = 10.2TGTDD470 pKa = 2.98HH471 pKa = 7.29TAFIDD476 pKa = 3.44EE477 pKa = 4.45WLDD480 pKa = 3.77SEE482 pKa = 4.71TTPEE486 pKa = 3.7
Molecular weight: 52.88 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A543AXL1|A0A543AXL1_9ACTN Acetyltransferase (GNAT) family protein OS=Stackebrandtia endophytica OX=1496996 GN=FB566_2872 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AIVAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84GRR40 pKa = 11.84ARR42 pKa = 11.84LTAA45 pKa = 4.29
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AIVAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84GRR40 pKa = 11.84ARR42 pKa = 11.84LTAA45 pKa = 4.29
Molecular weight: 5.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1637824 |
29 |
6712 |
315.1 |
34.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.123 ± 0.04 | 0.795 ± 0.01 |
6.639 ± 0.033 | 5.615 ± 0.034 |
2.918 ± 0.02 | 8.715 ± 0.027 |
2.236 ± 0.016 | 4.364 ± 0.023 |
1.969 ± 0.027 | 10.003 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.088 ± 0.015 | 2.159 ± 0.02 |
5.657 ± 0.035 | 2.879 ± 0.021 |
7.466 ± 0.042 | 5.684 ± 0.02 |
6.452 ± 0.032 | 8.534 ± 0.03 |
1.565 ± 0.015 | 2.137 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
